Abstract
Pancreatic cancer is the eighth-leading cause of cancer-associated mortality worldwide. To date, the cellular and molecular mechanisms associated with the invasion and metastasis of pancreatic cancer remain unclear. To examine these mechanisms, a microRNA (miRNA/miR) microarray with 1,965 genes was hybridized with labeled miRNA probes from invasive PC-1.0 and non-invasive PC-1 cells for molecular profiling analysis. In addition, reverse transcription quantitative-polymerase chain reaction (RT-qPCR) was utilized to validate the microarray results. Online miRNA target prediction algorithms online were used to predict the target genes of the differentially expressed miRNAs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) term enrichment analysis were performed for the potential targets of the differentially expressed miRNAs. The results demonstrated that 54 miRNAs were differentially expressed, of which 33 were upregulated and 21 were downregulated in the PC-1.0 cell line compared with the PC-1 cell line. A total of 6 upregulated miRNAs (miR-31, −34a, −181a, −181b, −193a-3p, and −193b) and 4 downregulated miRNAs (miR-221, −222, −484, and −502-3p) were selected from these 54 miRNAs and validated by RT-qPCR. The differentially expressed miRNAs were further validated by RT-qPCR in the human pancreatic cancer cell lines AsPC-1 (highly invasive) and CAPAN-2 (less invasive). The results revealed that 2 upregulated miRNAs (miR-34a and −193a-3p) and 4 downregulated miRNAs (miR-221, −222, −484, and −502-3p) exhibited a consistent expression pattern between the PC-1.0/PC-1 and AsPC-1/CAPAN-2 pancreatic cancer cells. The GO and KEGG enrichment analysis indicated that the mRNAs potentially targeted by miRNAs were involved in a range of biological functions. These results suggest that different miRNA expression profiles occur between highly and weakly invasive and metastatic pancreatic cancer cell lines, and may affect a variety of biological functions in pancreatic cancer.
Keywords: microRNA, microarray, pancreatic cancer, invasion, metastasis
Introduction
Pancreatic cancer is the eighth-leading cause of cancer-associated mortality worldwide. The high frequency of pancreatic cancer invasion and metastasis results in an extremely poor prognosis, and is one of the most defining characteristics of pancreatic cancer. The majority of patients are incurable at the time of diagnosis, with a median survival time of <1 year, and a 5-year survival rate of 6% for all stages (1,2).
To date, the cellular and molecular mechanisms of invasion and metastasis in pancreatic cancer are incompletely characterized. The identification of the factors associated with differences in the potential for tumor invasion and metastasis may provide useful information for the development of novel therapeutic methods to prevent these outcomes. A number of functional studies have demonstrated that microRNAs (miRNAs/miRs) serve important roles in biological processes that affect tumor progression, including cell differentiation, migration, invasion, metastasis and epithelial-to-mesenchymal transition (EMT) (3–5). miRNA expression profiling experiments have been performed regarding a number of different types of cancer and have identified a large number of aberrantly regulated miRNAs that may contribute to carcinogenesis by promoting the expression of proto-oncogenes or inhibiting the expression of tumor suppressor genes, including in pancreatic cancer (6–8).
To investigate the mechanisms of invasion and metastasis in pancreatic cancer, two hamster pancreatic cancer cell lines with different potentials for invasion and metastasis following intrapancreatic transplantation, i.e., PC-1, with a low potential, and PC-1.0, with a high potential, were previously established by Egami et al (9), from a pancreatic ductal carcinoma induced by N-nitrosobis (2-oxopropyl) amine (BOP) in a golden Syrian hamster (10).
In the present study, the differential expression of miRNA in the hamster pancreatic cancer cell lines was analyzed utilizing miRNA microarray technology, and verified via RT-qPCR. In addition, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) term enrichment analysis were applied to provide further evidence that the differentially expressed miRNAs were markers for invasion and metastasis in pancreatic cancer.
Materials and methods
Cell lines and cell culture
Two hamster pancreatic cancer cell lines were used, including the weakly invasive, rarely metastatic cell line PC-1, and the highly invasive and metastatic cell line PC-1.0. The PC-1 cell line was established from pancreatic ductal adenocarcinomas induced by BOP in a golden Syrian hamster (9). The PC-1.0 cell line was established from a subcutaneous tumor produced after the inoculation of PC-1 cells into hamsters (10). These two cell lines exhibit different growth rates and morphology in vitro: PC-1 cells form island-like cell colonies, whereas PC-1.0 cells primarily grow as single cells (11). The human pancreatic cancer cell lines AsPC-1 (highly invasive) and CAPAN-2 (less invasive) were also used. CAPAN-2 cells grow primarily as island-like colonies, similar to PC-1 cells, whereas AsPC-1 cells exhibit a growth pattern of single cells, similar to PC-1.0 cells. The PC-1.0 and PC-1 cells were given as a gift from Professor Baba H. The AsPC-1 and CAPAN-2 cell lines were purchased from the Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, China).
All cell lines were grown in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin G, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Preparation of total RNA
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After TRIzol extraction, RNA was further purified using an RNeasy mini spin column kit (Qiagen, Inc., Valencia, CA, USA). The concentration and quality of the RNA were assessed via spectrophotometry and agarose gel electrophoresis.
miRNA microarray
The miRNA microarray chip version 3.0 (CapitalBio Technology, Inc., Beijing, China) contained 1,965 mature miRNA probes; as the hamster gene sequence was not complete at the time of the study, the microarray chip used in the present study was designed as a mixed gene chip, including 988 human, 350 rat and 627 mouse miRNA genes. A total of 1,965 probes were designed on the basis of the sequences present in the miRBase version 12.0 miRNA database (12). These probes were labeled onto a 75×25 mm chemically-modified plate using the SmartArray™ microarray system (CapitalBio Technology, Inc.). The samples also contained two endogenous controls (U6, tRNA), eight exogenous controls (Zip5, Zip13, Zip15, Zip21, Zip23, Zip25, Y2 and Y3; Ambion; Thermo Fisher Scientific, Inc.), a positive control (HEX), and a hybridization negative control (50% dimethyl sulfoxide). The control sequences are listed in Table I.
Table I.
Control and normalization sequences for the microRNA microarray.
| Identity | Sequence (5′-3′) |
|---|---|
| U6 | ATTTGCGTGTCATCCTTGCG |
| tRNA | GGGTTATGGGCCCAGCACGCTTCCGCTGCGCCACTCTGCT |
| Zip23 | CAGCATCGGACCGGTAATCGGACC |
| Zip5 | GACCACCTTGCGATCGGGTACAGC |
| Zip15 | GACCGGTATGCGACCTGGTATGCG |
| Zip13 | CAGCGGTAGACCACCTATCGTGCG |
| Zip21 | TGCGATCGCAGCGGTAACCTGACC |
| Zip25 | GACCATAGTGCGGGTAGGTAGACC |
| Y2 | AGGTACGAAACGCTAAGAAT |
| Y3 | CATTCCTAAACGGGCTGAT |
| HEX | GTCACATGCGATGGATCGAGCTCCTTTATCATCGTTCCCACCTTAATGCA |
To isolate miRNA, total RNA (40.0 µg) was prepared using the polyethylene glycol (PEG) method; high molecular weight RNAs were removed by precipitation with 12.5% PEG-8000 and 1.25 M NaCl. The remaining RNA molecules were fractionated on a 15% acrylamide gel containing 8 M urea and extracted in water. Subsequently, the isolated miRNAs were dephosphorylated with calf intestinal alkaline phosphatase and labeled with CU-cy3 (green) and CU-cy5 (red; GE Healthcare Dharmacon, Inc., Lafayette, CO, USA), respectively, utilizing T4 RNA ligase to couple the 3′ end of the RNAs. The labeled products were isolated, purified and hybridized using a hybridization solution (15% formamide, 0.2% SDS, 3X SSC, 5X Denhardt's solution) at 42°C overnight. The plate was washed separately with solution I (0.2% SDS and 2X SSC) and solution II (0.2X SSC) for 4 min, dried, and scanned using a LuxScan 10K/A dual pathways laser scanner (CapitalBio Technology, Inc.).
Microarray analysis
miRNA profiles were adjusted with the global mean values to establish uniformity according to the total signal intensity of Cy5 and Cy3. The data were normalized and summarized using the LOWESS method, as previously described (13). The miRNAs were labeled according to the intensity of the signal and the quality of the image. Signal values >400 and <1,500 or >1,500 were selected. The two iterations of the microarray with different fluorescence labels were integrated as ratio=(ratio 1 × ratio 2)0.5 (Fig. 1). The most significant differentially expressed miRNAs (ratio ≥2 or ≤0.5, and q-value <1%) were identified following the integration. The miRNA probes tested the mature miRNA* and miRNA simultaneously, which originated from the same hairpin miR-precursors. The miRNA labeled with “*” represented a lower expression of miRNA when the miRNA* and miRNA were detected in the same cell line (Table II and III). Each miRNA gene was in the microarray in triplicate. The data were analyzed using Significance Analysis of Microarrays software (version 3.02) (14).
Figure 1.
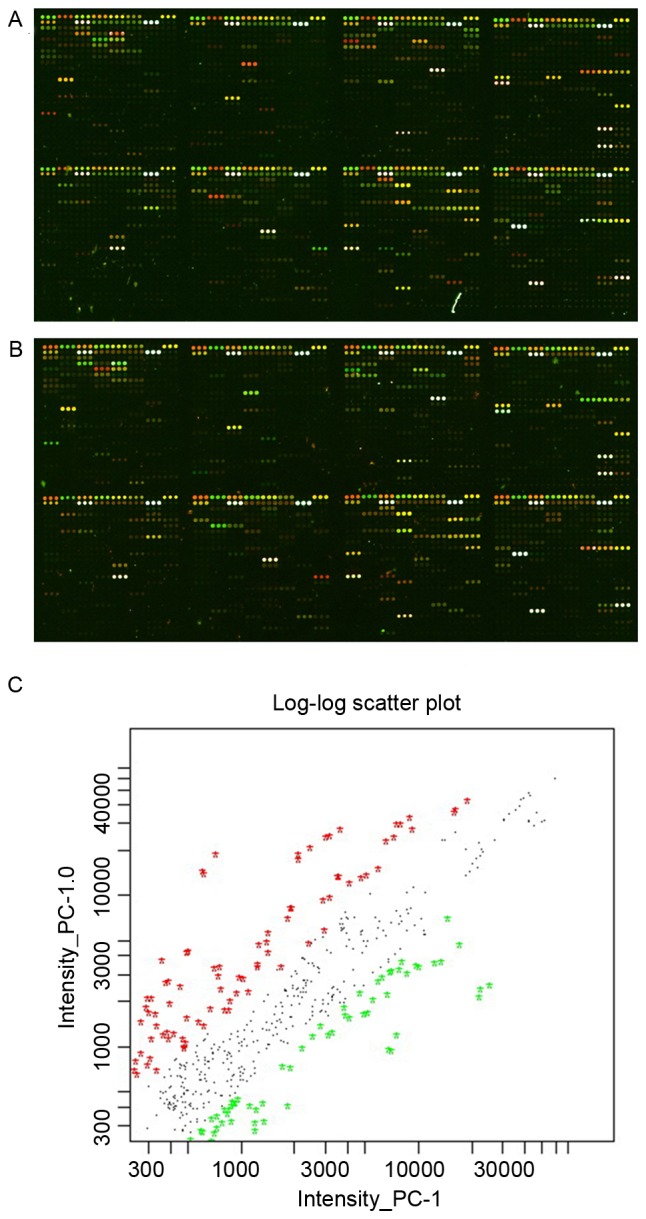
miRNA chip overlay images. (A) Original image of two-channel miRNA microarray; PC-1.0 was labeled with red fluorescent Cy5 dye and PC-1 was labeled with green fluorescent Cy3 dye. (B) Original image of two-channel miRNA microarray; PC-1.0 was labeled with green fluorescent Cy5 dye and PC-1 was labeled with red fluorescent Cy3 dye. (C) Scatter plot of PC-1.0 and PC-1 data. miRNA, microRNA.
Table II.
miRNAs upregulated in the highly invasive and metastatic cells (PC-1.0) compared with the weakly invasive and metastatic cells (PC-1).
| miRNA | Score (d) | q-value (%) |
|---|---|---|
| hsa-miR-181a | 20.95824611 | 0 |
| hsa-miR-486-3p | 5.0196028 | 0 |
| hsa-miR-31* | 13.35982897 | 0 |
| hsa-miR-181b | 12.89899491 | 0 |
| hsa-miR-31 | 23.80535775 | 0 |
| mmu-miR-193b | 39.5763408 | 0 |
| PREDICTED_miR229 | 9.955537815 | 0 |
| hsa-miR-193a-3p | 22.46659934 | 0 |
| hsa-miR-487b | 7.75232753 | 0 |
| hsa-miR-193b | 11.55311539 | 0 |
| hsa-miR-34a | 9.876005705 | 0 |
| hsa-miR-1538 | 5.385994942 | 0 |
| PREDICTED_miR145 | 4.44252184 | 0 |
| hsa-miR-708 | 23.71513905 | 0 |
| hsa-miR-146a | 8.050169358 | 0 |
| hsa-miR-128 | 5.398905767 | 0 |
| hsa-miR-1273 | 6.631137944 | 0 |
| hsa-miR-205 | 10.03459558 | 0 |
| hsa-miR-141 | 5.271252768 | 0 |
| hsa-miR-629* | 10.52079077 | 0 |
| hsa-miR-410 | 9.782045622 | 0 |
| hsa-miR-200a | 29.2232436 | 0 |
| rno-miR-25* | 9.273404082 | 0 |
| hsa-miR-1308 | 8.86116404 | 0 |
| hsa-let-7i* | 20.19289615 | 0 |
| hsa-miR-615-5p | 5.219655733 | 0 |
| hsa-miR-125b | 11.47936434 | 0 |
| hsa-miR-29b | 24.23706017 | 0 |
| hsa-miR-101 | 9.76085076 | 0 |
| hsa-miR-27a | 11.34655117 | 0 |
| mmu-miR-433* | 7.455889793 | 0 |
| hsa-miR-181c | 9.99770499 | 0 |
| hsa-let-7i | 18.34707892 | 0 |
Indicates miRNAs with low expression compared with the high expression of hairpin miR-precursors. hsa, Homo sapiens; miRNA/miR, microRNA; mmu, Mus musculus; rno, Rattus norvegicus.
Table III.
miRNAs downregulated in highly invasive and metastatic cells (PC-1.0) compared with weakly invasive and metastatic cells (PC-1).
| miRNA | Score (d) | q-value (%) |
|---|---|---|
| hsa-miR-324-3p | −7.617631275 | 0 |
| hsa-let-7d | −12.61068994 | 0 |
| hsa-miR-7 | −10.69123677 | 0 |
| mmu-miR-324-3p | −4.965056712 | 0 |
| hsa-let-7c | −8.811013746 | 0 |
| hsa-let-7a | −13.24229019 | 0 |
| hsa-miR-320b | −10.34928534 | 0 |
| rno-miR-204* | −6.090373954 | 0 |
| hsa-miR-107 | −5.951571765 | 0 |
| hsa-miR-500* | −5.384135129 | 0 |
| hsa-miR-378 | −19.88076516 | 0 |
| hsa-miR-30c | −21.64210903 | 0 |
| hsa-miR-378* | −6.871730972 | 0 |
| hsa-miR-186 | −5.551907546 | 0 |
| hsa-miR-221 | −28.19576008 | 0 |
| hsa-miR-484 | −13.78058027 | 0 |
| hsa-miR-502-3p | −11.19826264 | 0 |
| mmu-miR-298 | −8.830289897 | 0 |
| mmu-miR-500 | −4.229250653 | 0 |
| mmu-miR-706 | −22.3542206 | 0 |
| hsa-miR-222 | −40.14461092 | 0 |
Indicates miRNAs with low expression compared with the high expression of hairpin miR-precursors. hsa, Homo sapiens; miRNA/miR, microRNA; mmu, Mus musculus; rno, Rattus norvegicus.
Reverse transcription quantitative-polymerase chain reaction (RT-qPCR)
The miRNAs were extracted using the mirVana™ microRNA isolation kit (Ambion; Thermo Fisher Scientific, Inc.). The miRNA levels were determined using the TaqMan® MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cDNA was amplified using mature miRNA-specific RT primers and TaqMan® MiRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol.
qPCR was performed on an ABI 7500 Real-Time PCR system using TaqMan 2X Universal PCR Master Mix II and the 20X Small RNA Assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) with a total volume of 20 µl. The amplification reactions were performed in triplicate in a 96-well plate using the following cycle: 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. The Cq values were calculated using the ABI Sequence Detection System software version 2.1. The noncoding small nuclear RNA U6 primer (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used as the endogenous control. The relative fold change for each miRNA was calculated using the comparative Cq (2−ΔΔCq) method (15). The primer sequences are listed in Table IV.
Table IV.
Sequences used in reverse transcription-quantitative polymerase chain reaction.
| miRNA | Probe sequence (5′-3′) |
|---|---|
| hsa-miR-31 | CAGCTATGCCAGCATCTTGCCT |
| hsa-miR-34a | AACAACCAGCTAAGACACTGCCA |
| hsa-miR-181a | ACTCACCGACAGCGTTGAATGTT |
| hsa-miR-181b | CCCACCGACAGCAATGAATGTT |
| hsa-miR-193a-3p | CTGGGACTTTGTAGGCCAGTT |
| mmu-miR-193b | AGCGGGACTTTGTGGGCCAGTT |
| hsa-miR-221 | GAAACCCAGCAGACAATGTAGCT |
| hsa-miR-222 | ACCCAGTAGCCAGATGTAGCT |
| hsa-miR-502-3p | TGAATCCTTGCCCAGGTGCATT |
| hsa-miR-484 | ATCGGGAGGGGACTGAGCCTGA |
| U6 | GTGCTCGCTTCGGCAGCACATATACTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTT |
miRNA/miR, microRNA; hsa, Homo sapiens.
Prediction of the target genes of the miRNAs
The target genes of the miRNAs were predicted using miRWalk database v2.0 which integrated several softwares, including DIANAmT (http://diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi/), miRanda (http://www.microrna.org/microrna/home.do), miRDB (http://mirdb.org/miRDB/), miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), RNAhydrid (http://bibiserv.techfak.uni-bielefeld.de/rnahydrid/), PICTAR (http://pictar.mdc-berlin.de/), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html), RNA22 (http://cbcsrv.watson.ibm.com/rna22_targets.html), and Targetscan (http://www.targetscan.org). The target genes were designated as predicted downstream mRNAs by >6 softwares. Cytoscape software (version 3.0.0; www.cytoscape.org) was used to illustrate the relationships between miRNAs and predicted downstream genes (16).
GO analysis
GO analysis was performed to determine the main functions of the putative target genes of the differentially expressed miRNAs using the GO database (http://www.geneontology.org/). The analysis was carried out using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) with a Q-value statistical test for identifying significantly enriched terms; a final output of P≤0.05 was considered to indicate a statistically significant difference.
Pathway analysis
The putative target genes were analyzed using the KEGG pathway database (17) using DAVID software. Q≤0.05 was considered to represent a statistically significant difference. Cytoscape was used to illustrate the relationship between the miRNAs and KEGG terms.
Statistical analysis
The RT-qPCR data were assessed using an unpaired t-test in SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a significant difference. The Benjamini-Hochberg method was used to adjust the P-values from the GO and KEGG enrichment analyses.
Results
Differentially expressed miRNAs identified by miRNA microarray between PC-1.0 and PC-1 cells
Of the 1,965 mature miRNAs analyzed in the microarray experiments, 54 were determined to be differentially expressed. Of these, 33 (61.1%) were upregulated in the highly invasive and metastatic cells (PC-1.0) compared with the weakly invasive and metastatic cells (PC-1; Table II), whereas 21 (38.9%) were significantly downregulated (Table III).
Validation of miRNA expression in the PC-1.0 and PC-1 hamster pancreatic cancer cells using RT-qPCR
To determine the reliability of the miRNA microarray data, 6 up-regulated miRNAs (miR-31, −34a, −181a, −181b, −193a-3p and −193b) and 4 down-regulated miRNAs (miR-221, −222, −484 and −502-3p), which varied significantly between the PC-1.0 and PC-1 cell lines in the microarray, were selected to be verified by RT-qPCR. The results were similar to those obtained using the miRNA microarray data, supporting the reliability of the expression data (Fig. 2).
Figure 2.
Reverse transcription-quantitative polymerase chain reaction analysis of highly (PC-1.0) and weakly (PC-1) invasive and metastatic hamster pancreatic cancer cells. A total of 10 miRNAs were selected to verify the reliability of the miRNA microarray data. Of the 10 miRNAs, 6 were upregulated and 4 were downregulated. These results were very similar to the miRNA microarray data, supporting its reliability. Black bar, PC-1.0 cell line. White bar, PC-1 cell line. *P<0.05. miRNA, microRNA.
Validation miRNA expression in the AsPC-1 and CAPAN-2 human pancreatic cancer cells using RT-qPCR
The results from the hamster pancreatic cancer cells were different from those in human cancer cells. A total of 6 of the 10 miRNAs had the same expression tendency in the PC-1.0/PC-1 and AsPC-1/CAPAN-2 pancreatic cancer cell lines, including miR-34a, −193a (upregulated), −221, −222, −484 and −502-3p (downregulated; Fig. 3).
Figure 3.
Reverse transcription-quantitative polymerase chain reaction analysis of highly (AsPC-1) and weakly (CAPAN-2) invasive and metastatic human pancreatic cancer cells. To further investigate the association of miRNAs with invasion and metastasis in pancreatic cancer, two human pancreatic cancer cell lines were analyzed. The results were partially different from those in the PC-1.0/PC-1 cells; of the 10 miRNAs, 2 were upregulated and 8 were downregulated. Black bar, AsPC-1 cell line. White bar, CAPAN-2 cell line. *P<0.05. miRNA, microRNA.
Prediction of the target genes of the miRNAs
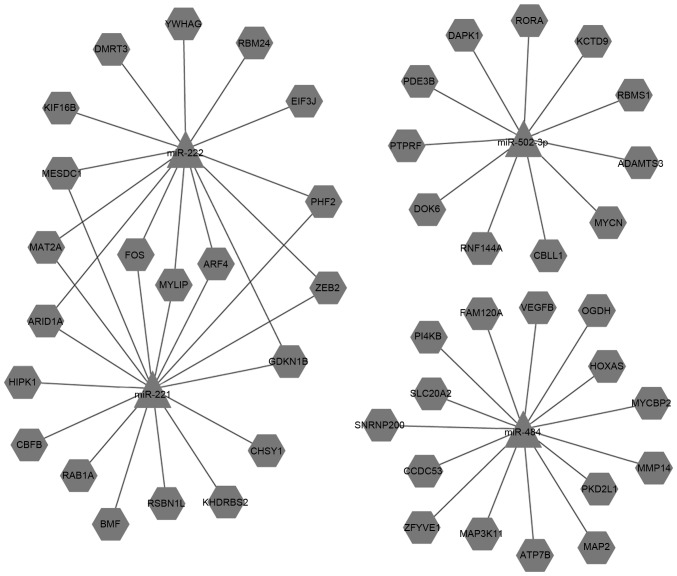
Various bioinformatic, experimental and combined approaches have been used to identify putative target genes for miRNAs; several databases that used these approaches applied in this study. There were 8,279 intersected target genes for miR-34a, 5,206 intersected target genes for miR-193a-3p, 5,990 intersected target genes for miR-221, 5,942 intersected target genes for miR-222, 8,722 intersected target genes for miR-484 and 4,582 intersected target genes for miR-502-3p. Selected important target genes (including upregulated and downregulated) are listed in Tables V and VI. Cytoscape software was used to illustrate the connections between the miRNAs and target genes (Figs. 4 and 5).
Table V.
Predicted target genes of upregulated miRNAs.
| miRNA | Target gene | Representative transcript | Gene name |
|---|---|---|---|
| miR-34a | NAV3 | NM_014903 | Neuron navigator 3 |
| ACSL1 | NM_001286711 | Acyl-CoA synthetase long-chain family member 1 | |
| AKAP6 | NM_004274 | A kinase (PRKA) anchor protein 6 | |
| CAPN6 | NM_014289 | Calpain 6 | |
| CORO1C | NM_014325 | Coronin, actin binding protein, 1C | |
| CTNND2 | NM_001288717 | Catenin (cadherin-associated protein), delta 2 | |
| E2F5 | NM_001951 | E2F transcription factor 5, p130-binding | |
| EML5 | NM_183387 | Echinoderm microtubule associated protein like 5 | |
| JAG1 | NM_000214 | Jagged 1 | |
| KIAA1217 | NM_001098500 | KIAA1217 | |
| LEF1 | NM_001130714 | Lymphoid enhancer-binding factor 1 | |
| LGR4 | NM_018490 | Leucine-rich repeat containing G protein-coupled receptor 4 | |
| MAP2K1 | NM_002755 | Mitogen-activated protein kinase kinase 1 | |
| NOTCH1 | NM_017617 | Notch 1 | |
| PDGFRA | NM_006206 | Platelet-derived growth factor receptor, alpha polypeptide | |
| PNOC | NM_006228 | Prepronociceptin | |
| TMEM55A | NM_018710 | Transmembrane protein 55A | |
| UHRF2 | NM_152896 | Ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase | |
| ZDHHC17 | NM_015336 | Zinc finger, DHHC-type containing 17 | |
| ZNF281 | NM_012482 | Zinc finger protein 281 | |
| miR-193a-3p | DCAF7 | NM_001003725 | DDB1 and CUL4 associated factor 7 |
| TMEM30A | NM_001143958 | Transmembrane protein 30A | |
| KCNJ2 | NM_000891 | Potassium channel, inwardly rectifying subfamily J, member 2 | |
| HOXD13 | NM_000523 | Homeobox D13 | |
| FHDC1 | NM_033393 | FH2 domain containing 1 | |
| EN2 | NM_001427 | Engrailed homeobox 2 | |
| DNAJC13 | NM_015268 | DnaJ (Hsp40) homolog, subfamily C, member 13 | |
| CTDSPL2 | NM_016396 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 | |
| CNOT6 | NM_015455 | CCR4-NOT transcription complex, subunit 6 | |
| CALB1 | NM_001740 | Calbindin 1, 28 kDa | |
| KRAS | NM_004985 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | |
| PLAU | NM_001145031 | Plasminogen activator, urokinase | |
| MMP19 | NM_002429 | Matrix metallopeptidase 19 | |
| JMY | NM_152405 | Junction mediating and regulatory protein, p53 cofactor | |
| MAPK8 | NM_001278547 | Mitogen-activated protein kinase 8 | |
| MAX | NM_002382 | MYC associated factor X |
miRNA/miR, microRNA.
Table VI.
Predicted target genes of downregulated miRNAs.
| miRNA | Target gene | Representative transcript | Gene name |
|---|---|---|---|
| miR-221 | KHDRBS2 | NM_152688 | KH domain containing, RNA binding, signal transduction associated 2 |
| FOS | NM_005252 | FBJ murine osteosarcoma viral oncogene homolog | |
| ARID1A | NM_006015 | AT rich interactive domain 1A (SWI-like) | |
| BMF | NM_001003943 | Bcl2 modifying factor | |
| HIPK1 | NM_181358 | Homeodomain interacting protein kinase 1 | |
| MESDC1 | NM_022566 | Mesoderm development candidate 1 | |
| MAT2A | NM_005911 | Methionine adenosyltransferase II, alpha | |
| ZEB2 | NM_001171653 | Zinc finger E-box binding homeobox 2 | |
| MYLIP | NM_013262 | Myosin regulatory light chain interacting protein | |
| PHF2 | NM_005392 | PHD finger protein 2 | |
| RSBN1L | NM_198467 | Round spermatid basic protein 1-like | |
| ARF4 | NM_001660 | ADP-ribosylation factor 4 | |
| CBFB | NM_001755 | Core-binding factor, beta subunit | |
| CDKN1B | NM_004064 | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | |
| CHSY1 | NM_014918 | Chondroitin sulfate synthase 1 | |
| RAB1A | NM_004161 | RAB1A, member RAS oncogene family | |
| miR-222 | ARF4 | NM_001660 | ADP-ribosylation factor 4 |
| ARID1A | NM_006015 | AT rich interactive domain 1A (SWI-like) | |
| CDKN1B | NM_004064 | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | |
| DMRT3 | NM_021240 | Doublesex and mab-3 related transcription factor 3 | |
| EIF3J | NM_001284335 | Eukaryotic translation initiation factor 3, subunit J | |
| FOS | NM_005252 | FBJ murine osteosarcoma viral oncogene homolog | |
| KIF16B | NM_001199865 | Kinesin family member 16B | |
| MAT2A | NM_005911 | Methionine adenosyltransferase II, alpha | |
| MESDC1 | NM_022566 | Mesoderm development candidate 1 | |
| MYLIP | NM_013262 | Myosin regulatory light chain interacting protein | |
| PHF2 | NM_005392 | PHD finger protein 2 | |
| RBM24 | NM_001143941 | RNA binding motif protein 24 | |
| YWHAG | NM_012479 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma | |
| ZEB2 | NM_001171653 | Zinc finger E-box binding homeobox 2 | |
| miR-484 | SNRNP200 | NM_014014 | Small nuclear ribonucleoprotein 200 kDa (U5) |
| CCDC53 | NM_001301107 | Coiled-coil domain containing 53 | |
| FAM120A | NM_001286722 | Family with sequence similarity 120A | |
| HOXA5 | NM_019102 | Homeobox A5 | |
| MAP2 | NM_002374 | Microtubule-associated protein 2 | |
| OGDH | NM_002541 | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | |
| PKD2L1 | NM_016112 | Polycystic kidney disease 2-like 1 | |
| SLC20A2 | NM_006749 | Solute carrier family 20 (phosphate transporter), member 2 | |
| VEGFB | NM_003377 | Vascular endothelial growth factor B | |
| ZFYVE1 | NM_021260 | Zinc finger, FYVE domain containing 1 | |
| MAP3K11 | NM_002419 | Mitogen-activated protein kinase kinase kinase 11 | |
| PI4KB | NM_001198773 | Phosphatidylinositol 4-kinase, catalytic, beta | |
| ATP7B | NM_000053 | ATPase, Cu++ transporting, beta polypeptide | |
| MYCBP2 | NM_015057 | MYC binding protein 2 | |
| MMP14 | NM_004995 | Matrix metallopeptidase 14 (membrane-inserted) | |
| miR-502-3p | KCTD9 | NM_017634 | Potassium channel tetramerisation domain containing 9 |
| RNF144A | NM_014746 | Ring finger protein 144A | |
| DOK6 | NM_152721 | Docking protein 6 | |
| PTPRF | NM_002840 | Protein tyrosine phosphatase, receptor type, F | |
| PDE3B | NM_000922 | Phosphodiesterase 3B, cGMP-inhibited | |
| RORA | NM_002943 | RAR-related orphan receptor A | |
| MYCN | NM_005378 | V-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog | |
| DAPK1 | NM_004938 | Death-associated protein kinase 1 | |
| ADAMTS3 | NM_014243 | ADAM metallopeptidase with thrombospondin type 1 motif, 3 | |
| CBLL1 | NM_024814 | Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase | |
| RBMS1 | NM_002897 | RNA binding motif, single stranded interacting protein 1 |
miRNA/miR, microRNA.
Figure 4.
Downstream target genes of the upregulated microRNAs.
Figure 5.
Downstream target genes of the downregulated microRNAs.
Gene ontology enrichment analysis
To understand the biological functions of the differently expressed miRNAs in different cellular processes, a GO enrichment analysis was performed using DAVID software, including the cellular component, molecular function and biological process categories. The upregulated and downregulated miRNAs were analyzed separately.
A total of 254 cellular component terms were enriched in the upregulated miRNAs and 273 in the downregulated miRNAs. Several of the terms were common between upregulated and downregulated miRNAs, including ‘nucleus’, ‘cytoplasm’, ‘membrane’, ‘extracellular region’, ‘Golgi apparatus’, ‘cytosol’, ‘endoplasmic reticulum’, ‘cytoskeleton’, ‘cell junction’, and ‘mitochondria’. ‘Nucleoplasm’ and ‘microtubules’ were more enriched in the upregulated miRNAs than the downregulated miRNAs. ‘Cell-cell adherens junctions’ was particularly associated with the upregulated miRNAs, whereas ‘tight junctions’ was associated with the downregulated miRNAs (Fig. 6).
Figure 6.
Cellular components for the differentially expressed microRNAs identified with Gene Ontology term enrichment analysis.
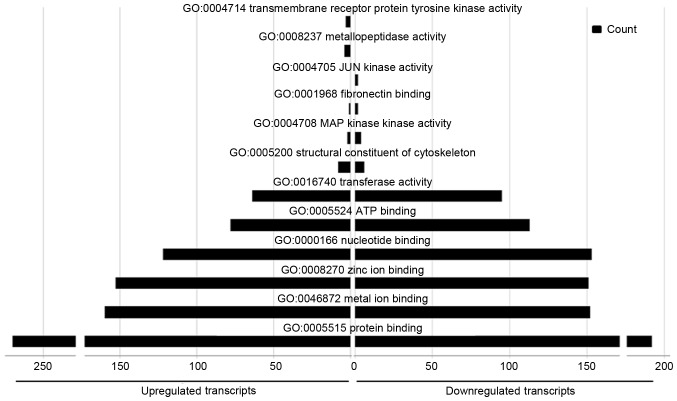
A total of 528 GO molecular function terms were enriched in the upregulated miRNAs, and 583 in the downregulated miRNAs. Several of the terms were common between sets, including ‘protein binding’, ‘metal ion binding’, ‘zinc ion binding’, ‘nucleotide binding’, ‘ATP binding’ and ‘transferase activity’. Several functions were particularly enriched in the upregulated miRNA set, including ‘structural constituents of the cytoskeleton’, whereas ‘MAP kinase kinase activity’ and ‘fibronectin-binding activity’ were more representative of the downregulated miRNAs. ‘Tyrosine kinase activity’ and ‘metallopeptidase activity in transmembrane receptor proteins’ were particularly represented in the upregulated miRNAs, and ‘JUN kinase activity’ was particularly represented in the downregulated miRNAs (Fig. 7).
Figure 7.
Molecular functions for the differentially expressed microRNAs identified with Gene Ontology term enrichment analysis.
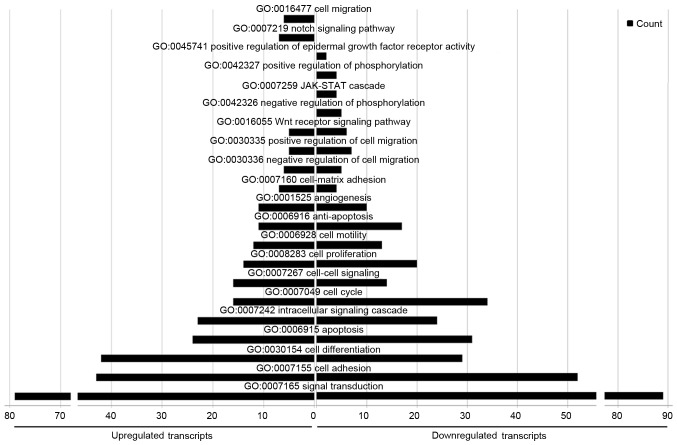
With regard to GO biological processes, 1,021 terms were enriched in the upregulated miRNAs and 1,280 in the downregulated miRNAs. As for the cellular component and molecular function categories, several biological processes were in common between the groups, including ‘signal transduction’, ‘cell adhesion’, ‘apoptosis’, ‘cell proliferation’, ‘cell motility’, ‘anti-apoptosis’, ‘angiogenesis’, ‘positive regulation of cell migration’ and ‘Wnt receptor signaling’. However, ‘cell-matrix adhesion’, ‘cell-cell signaling’, and ‘cell differentiation’ were more enriched in the up-regulated miRNAs than the downregulated miRNAs, whereas ‘cell cycle processes’ was more enriched in the downregulated miRNAs. In particular, ‘cell migration’ and ‘Notch signaling pathways’ were only represented in the upregulated miRNAs, whereas ‘positive regulation of epidermal growth factor receptor activity’, ‘positive regulation of phosphorylation’, ‘JAK-STAT pathway’ and ‘negative regulation of phosphorylation’ were only represented in the downregulated miRNAs (Fig. 8).
Figure 8.
Biological processes for the differentially expressed microRNAs identified with Gene Ontology term enrichment analysis.
KEGG enrichment analysis
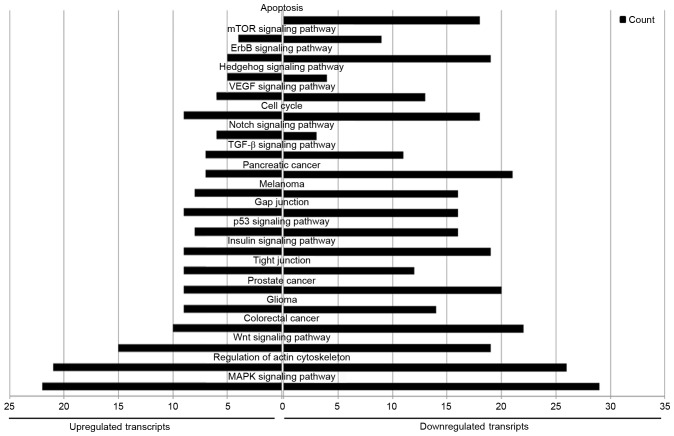
KEGG is a database of genetic and molecular networks. A total of 91 pathways were associated with the upregulated miRNAs, and 112 with the downregulated miRNAs. There were 74 pathways in common between the upregulated and downregulated miRNAs, including the ‘MAPK signaling pathway’, ‘regulation of actin cytoskeleton’, ‘Wnt signaling pathway’, ‘pancreatic cancer’, ‘colorectal cancer’, ‘tight junctions’, ‘p53 signaling pathway’, ‘gap junctions’, ‘TGF-beta signaling pathway’, ‘Notch signaling pathway’, ‘cell cycle’ and ‘mTOR signaling pathway’. Furthermore, 16 pathways were associated with only the upregulated miRNAs, and 36 with only the downregulated miRNAs. ‘Apoptosis pathway’ was particularly enriched in the downregulated miRNAs (Fig. 9). Cytoscape was used to illustrate the connections between the miRNAs and pathways (Fig. 10).
Figure 9.
Kyoto Encyclopedia of Genes and Genomes term enrichment analysis.
Figure 10.
Kyoto Encyclopedia of Genes and Genomes term pathways common or unique to the up- and downregulated microRNAs.
Discussion
The highly (PC-1.0) and weakly (PC-1) invasive and metastatic pancreatic cancer cell lines, which were established from an experimental pancreatic cancer model a previous study by Egami et al (9,10), exhibit clearly different potentials for invasion and metastasis (11). To further investigate the mechanisms of the invasion and metastasis of pancreatic cancer in the present study, highly (AsPC-1) and weakly (CAPAN-2) invasive and metastatic cell lines were selected, as they possess similar biological characteristics to the PC-1.0/PC-1 cell lines when compared with other human pancreatic cancer cell lines such as CAPAN-1 or MiaPACA-2. Many factors have been identified that are involved in the mechanisms of invasion and metastasis in both hamster and human pancreatic cancer cell lines, including the tight junction factors [claudin-1 (18), ZO-1 (19) and occludin (20)], MMP-7 (21,22) and mitogen-activated protein kinase (MAPK) signaling pathway factors [ERK1/2 (23), MEK2 (24) and EGFR (25)]. We hypothesize that the mechanisms and key factors of PC-1.0/PC-1 cells are similarly expressed and serve a vital role in human pancreatic cancer cells, with the same biological functions.
The Syrian hamster has been verified as a unique model for investigating pancreatic cancer by Pour et al (26). Hamster and human genes have a high similarity (27), which may explain why the RT-qPCR results in human cells were similar to those of the hamster cells. Since the hamster genome sequence was not complete at the time of the study, the microarray chip in the present study was designed as a mixed gene chip, including human, rat and mouse genes. miRNAs with high similarity scores were selected for use in the array experiments, with a focus on human miRNAs, as the ultimate goal was to investigate the mechanisms in humans. To verify the results, RT-qPCR was used to analyze the hamster and human pancreatic cancer cells, with similar results being identified. The PC-1.0 and PC-1 cells were more homologous than the AsPC-1 and CAPAN-2 cell lines, hence why they were selected for microarray analysis instead of the human cell lines. The differentially expressed miRNAs obtained from the PC-1.0 and PC-1 cell lines were validated by RT-qPCR using AsPC-1 and CAPAN-2. It is hoped that the final results of this analysis will contribute to developing novel approaches for clinical therapy.
A total of 2 upregulated miRNAs (miR-34a and −193a-3p) and 4 downregulated miRNAs (miR-221, −222, −484, and −502-3p) were selected and examined between the PC-1.0/PC-1 and AsPC-1/CAPAN-2 cell lines in the present study. The results indicated that miR-34a and −193a-3p may promote the progression of invasion and metastasis in pancreatic cancer, whereas miR-221, −222, −484 and −502-3p may prevent this. To date, several studies have evaluated invasion and metastasis in pancreatic cancers (28–31); however, only a few studies reported data regarding the miRNAs identified in the present study. miR-34a is a highly conserved miRNA that is known to be a downstream target of p53, and a tumor suppressor (32). Yang et al (33) observed that miR-34a was significantly upregulated in uveal melanoma via a miRNA microarray. Lee et al (34) reported that miR-222 was upregulated in pancreatic cancer tissue compared with adjacent normal tissue, and was associated with cell proliferation. In addition, miR-221 was reported to be upregulated in pancreatic cancer tissues, cell lines and pre-operative patient blood plasma, and downregulated following surgery (35). This result was in contrast with the present study. Therefore, more study will be required to evaluate the potential of differentially expressed miRNAs as markers of invasion and metastasis in pancreatic cancer.
The mechanisms associated with invasion and metastasis in pancreatic cancer are complex and incompletely elucidated. In the present study, GO term and KEGG pathway enrichment analyses were used to investigate the differences in the biological functions of highly and weakly invasive and metastatic pancreatic cancer cell lines. The upregulated miRNAs were primarily associated with ‘cell-cell adherens junctions’, ‘metallopeptidase activity’, ‘cell migration’ and ‘Notch signaling pathway’, whereas the downregulated miRNAs were associated primarily with ‘tight junctions’, ‘JAK-STAT pathway’ and ‘apoptosis’. The overlap between the up- and downregulated miRNAs may indicate the presence of intricate cross-talk in the regulation of pancreatic cancer. ‘MAP kinase kinase activity’, for example, was enriched in both up- and downregulated miRNAs. In a previous study, Tan et al (20) demonstrated that MMP-7 was associated with cell dissociation, forming a positive feedback loop with the activation of the epidermal growth factor receptor-mediated MAPK signaling pathway. In the present study, KEGG analysis indicated that ‘apoptosis’ was predominantly enriched in the downregulated miRNAs. Therefore, we hypothesize that the upregulated miRNAs miR-34a and −193a-3p may be primarily involved in cell-cell adherens junctions, metallopeptidase activity and cell migration, whereas the downregulated miRNAs miR-221, −222, −484 and −502-3p may be primarily associated with tight junctions and apoptosis in pancreatic cancer cell lines.
In conclusion, these results suggest that distinct miRNA expression profiles occur between highly and weakly invasive and metastatic pancreatic cancer cell lines. In addition, differentially expressed miRNAs may be involved in a variety of biological functions and mechanisms in pancreatic cancer. In this context, the identification of invasive and metastatic-specific miRNAs may allow the development of novel therapeutic and diagnostic strategies to target invasion and metastasis in pancreatic cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant-in-aid from the National Nature Science Foundation of China (grant no., 30973501).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author, on reasonable request.
Authors' contributions
XT designed the experiments and was responsible for the quality control of the data. LZ performed the miRNA microarray, interpreted the data and was the main contributor in writing the manuscript. YS and YY maintained the cell lines and prepared the total RNA. HL and HW performed the RT-qPCR. ZW predicted the target genes of the miRNAs. XZ and FG performed GO and KEGG analysis. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Bandres E, Agirre X, Ramirez N, Zarate R, Garcia-Foncillas J. MicroRNAs as cancer players: Potential clinical and biological effects. DNA Cell Biol. 2007;26:273–282. doi: 10.1089/dna.2006.0544. [DOI] [PubMed] [Google Scholar]
- 4.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs-the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 7.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets; Proc Natl Acad Sci USA; 2006; pp. 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Egami H, Takiyama Y, Cano M, Houser WH, Pour PM. Establishment of hamster pancreatic ductal carcinoma cell line (PC-1) producing blood group-related antigens. Carcinogenesis. 1989;10:861–869. doi: 10.1093/carcin/10.5.861. [DOI] [PubMed] [Google Scholar]
- 10.Egami H, Tomioka T, Tempero M, Kay D, Pour PM. Development of intrapancreatic transplantable model of pancreatic duct adenocarcinoma in Syrian golden hamsters. Am J Pathol. 1991;138:557–561. [PMC free article] [PubMed] [Google Scholar]
- 11.Pour PM, Egami H, Takiyama Y. Patterns of growth and metastases of induced pancreatic cancer in relation to the prognosis and its clinical implications. Gastroenterology. 1991;100:529–536. doi: 10.1016/0016-5085(91)90226-B. [DOI] [PubMed] [Google Scholar]
- 12.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database Issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response; Proc Natl Acad Sci USA; 2001; pp. 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan X, Egami H, Ishikawa S, Kurizaki T, Nakagawa M, Hirota M, Ogawa M. Arrangement of expression and distribution of tight junction protein claudin-1 in cell dissociation of pancreatic cancer cells. Int J Oncol. 2004;25:1567–1574. [PubMed] [Google Scholar]
- 19.Tan X, Egami H, Ishikawa S, Kurizaki T, Hirota M, Ogawa M. Zonula occludens-1 (ZO-1) redistribution is involved in the regulation of cell dissociation in pancreatic cancer cells. Dig Dis Sci. 2005;50:1402–1409. doi: 10.1007/s10620-005-2853-9. [DOI] [PubMed] [Google Scholar]
- 20.Tan X, Tamori Y, Egami H, Ishikawa S, Kurizaki T, Takai E, Hirota M, Ogawa M. Analysis of invasion-metastasis mechanism in pancreatic cancer: Involvement of tight junction transmembrane protein occludin and MEK/ERK signal transduction pathway in cancer cell dissociation. Oncol Rep. 2004;11:993–998. [PubMed] [Google Scholar]
- 21.Tan X, Egami H, Abe M, Nozawa F, Hirota M, Ogawa M. Involvement of MMP-7 in invasion of pancreatic cancer cells through activation of the EGFR mediated MEK-ERK signal transduction pathway. J Clin Pathol. 2005;58:1242–1248. doi: 10.1136/jcp.2004.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan X, Egami H, Ishikawa S, Sugita H, Kamohara H, Nakagawa M, Nozawa F, Abe M, Ogawa M. Involvement of matrix metalloproteinase-7 in invasion-metastasis through induction of cell dissociation in pancreatic cancer. Int J Oncol. 2005;26:1283–1289. [PubMed] [Google Scholar]
- 23.Tan X, Egami H, Ishikawa S, Kurizaki T, Tamori Y, Takai E, Hirota M, Ogawa M. Relationship between the expression of extracellular signal-regulated kinase 1/2 and the dissociation of pancreatic cancer cells: Involvement of ERK1/2 in the dissociation status of cancer cells. Int J Oncol. 2004;24:815–820. [PubMed] [Google Scholar]
- 24.Tan X, Egami H, Kamohara H, Ishikawa S, Kurizaki T, Yoshida N, Tamori Y, Takai E, Hirota M, Ogawa M. Involvement of the mitogen-activated protein kinase kinase 2 in the induction of cell dissociation in pancreatic cancer. Int J Oncol. 2004;24:65–73. [PubMed] [Google Scholar]
- 25.Tan X, Egami H, Ishikawa S, Nakagawa M, Ishiko T, Kamohara H, Hirota M, Ogawa M. Relationship between activation of epidermal growth factor receptor and cell dissociation in pancreatic cancer. Int J Oncol. 2004;25:1303–1309. [PubMed] [Google Scholar]
- 26.Pour PM, Runge RG, Birt D, Gingell R, Lawson T, Nagel D, Wallcave L, Salmasi SZ. Current knowledge of pancreatic carcinogenesis in the hamster and its relevance to the human disease. Cancer. 1981;47(6 Suppl):S1573–S1589. doi: 10.1002/1097-0142(19810315)47:6+<1573::AID-CNCR2820471420>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa S, Egami H, Kurizaki T, Akagi J, Tamori Y, Yoshida N, Tan X, Hayashi N, Ogawa M. Identification of genes related to invasion and metastasis in pancreatic cancer by cDNA representational difference analysis. J Exp Clin Cancer Res. 2003;22:299–306. [PubMed] [Google Scholar]
- 28.Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, Tian K, Li X, Zhu S, Niu Y, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther. 2013;12:2569–2580. doi: 10.1158/1535-7163.MCT-13-0296. [DOI] [PubMed] [Google Scholar]
- 29.Kadera BE, Li L, Toste PA, Wu N, Adams C, Dawson DW, Donahue TR. MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis. PLoS One. 2013;8:e71978. doi: 10.1371/journal.pone.0071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. doi: 10.1186/1475-2867-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y, Wang B, Tian K, Deng SC, Li X, Zhu S, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One. 2013;8:e73803. doi: 10.1371/journal.pone.0073803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C, Wei W. The miRNA expression profile of the uveal melanoma. Sci China Life Sci. 2011;54:351–358. doi: 10.1007/s11427-011-4149-y. [DOI] [PubMed] [Google Scholar]
- 34.Lee C, He H, Jiang Y, Di Y, Yang F, Li J, Jin C, Fu D. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Med Oncol. 2013;30:700. doi: 10.1007/s12032-013-0700-y. [DOI] [PubMed] [Google Scholar]
- 35.Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author, on reasonable request.