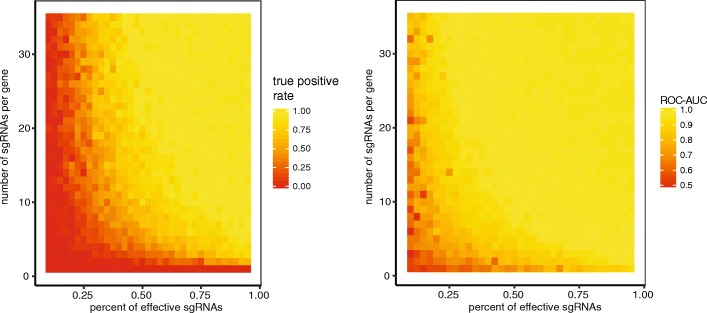
Fig. 4.
The true positive rate (the percentage of true interesting genes correctly identified) for CRISPhieRmix at an FDR of 0.1 (left) and ROC-AUC (right) for simulated data with varying percent of effective sgRNAs and number of sgRNAs per gene. The ROC-AUC is high for most parameters, implying that ranking the genes is usually easy, and the percent of correctly identified genes is much smaller, implying that it is difficult to correctly identify all truly interesting genes while simultaneously effectively controlling the false discovery rate