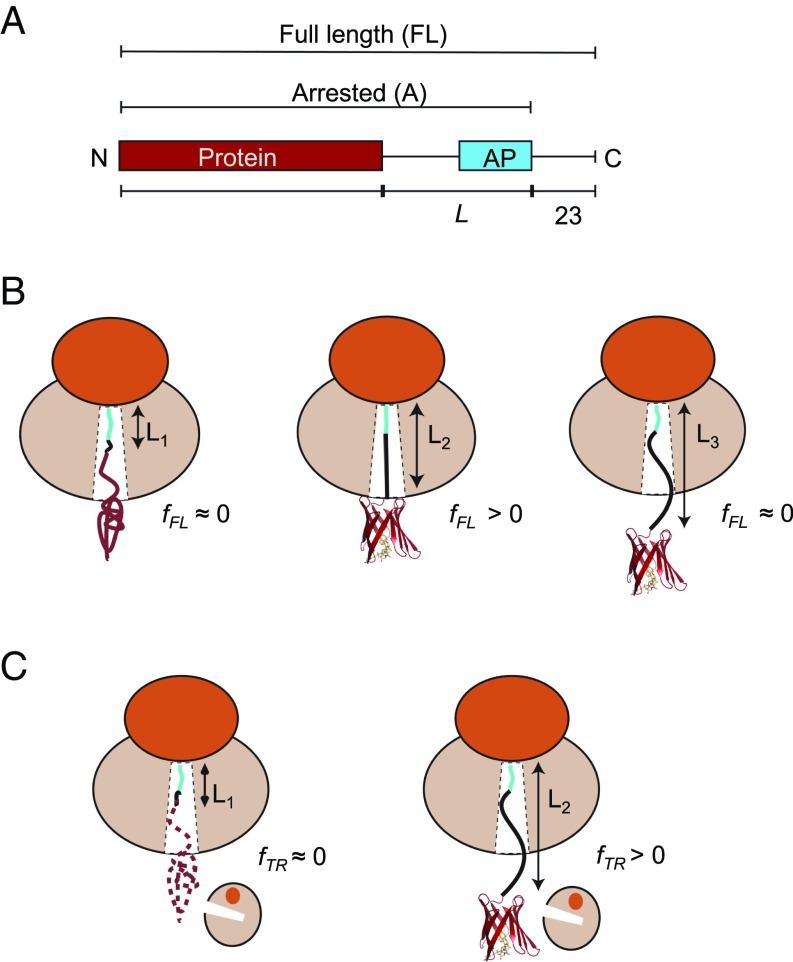
Fig. 1.
(A) Schematic representation of in vitro translated constructs. The protein under study is placed L residues upstream of the last proline in the AP, and a C-terminal 23-residue extension is added downstream of the AP to ensure that the arrested and full-length forms of the construct can be cleanly separated by SDS/PAGE. (B) The AP-based assay: At linker length L1, the protein is too deep in the exit tunnel to be able to fold, and hence no force is generated on the AP and mostly arrested nascent chains are produced (fFL ≈ 0). At L2, if the linker is stretched beyond its equilibrium length the protein can just reach a location in the exit tunnel where there is sufficient space for it to fold. Some of the folding free energy is therefore stored as elastic energy in the linker, increasing the force on the AP. More full-length protein is produced (fFL > 0). At L3, finally, the protein is already folded when the ribosome reaches the C-terminal end of the AP, and little force is exerted on the AP (fFL ≈ 0). (C) The on-ribosome pulse proteolysis assay: At L1, the protein is located too deep in the exit tunnel to be able to fold and the nascent chain is hence degraded by a brief thermolysin pulse. At L2, the protein is folded and hence resistant to proteolysis. Note that the protein is attached to the ribosome via a (GS)n linker that in itself is insensitive to thermolysin.