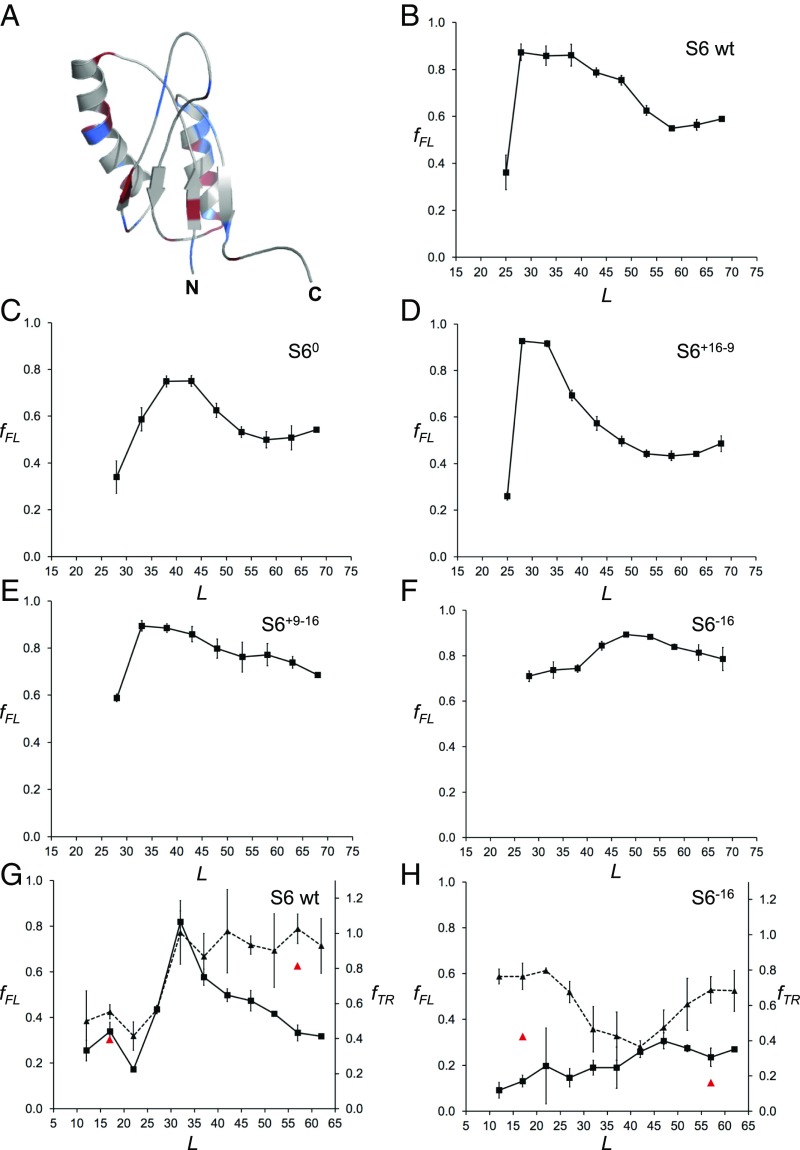
Fig. 4.
(A) NMR structure of wild-type S6 from T. thermophilus (PDB ID code 2kjv). Positively charged residues (Arg+Lys) are colored in blue and negatively charged residues (Asp+Glu) in red. (B) fFL profile of wild-type S6+16–16 (for sequences see SI Appendix, Fig. S2). (C) fFL profile of S60. (D) fFL profile of S6+16–9. (E) fFL profile of S6+9–16. (F) fFL profile of S6−16. (G) fFL (squares, solid curve) and fTR (triangles, dashed curve) profiles for wild-type S6. Red data points at L = 17 and 57 residues are for the destabilizing mutation L10A. (H) fFL (squares, solid curve) and fTR (triangles, dashed curve) profiles for S6−16. Red data points at L = 17 and 57 residues are for the destabilizing mutation L10A. The SecM(Ec) AP was used in A–F and the SecM(Ms) AP in G and H.