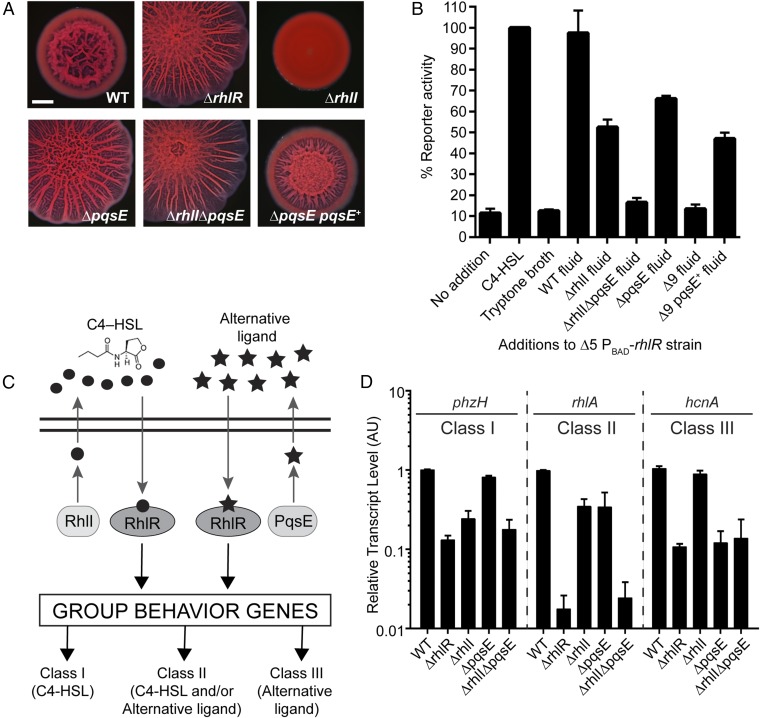
Fig. 1.
Identification of pqsE as the alternative-ligand synthase. (A) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 5 d of growth. pqsE+ refers to complementation with pqsE under the Plac promoter on the pUCP18 plasmid. (Scale bar, 2 mm.) (B) rhlA expression was measured using a chromosomally encoded PrhlA-mNeonGreen transcriptional reporter. The PA14 strain used in this analysis is ΔlasR ΔlasI ΔrhlR ΔrhlI ΔpqsE carrying PBAD-rhlR on the chromosome; designated Δ5 PBAD-rhlR. rhlR was induced with 0.1% l-arabinose. The rhlA reporter activity was set to 100% when 10 µM C4-HSL was added (second bar). For all other bars, either 30% (vol/vol) of broth or the indicated cell-free culture fluid was added. The bar designated Δ9 refers to culture fluid from the ΔlasR ΔlasI ΔrhlR ΔrhlI ΔpqsABCDE strain. The bar designated Δ9 pqsE+ refers to culture fluid from the ΔlasR ΔlasI ΔrhlR ΔrhlI ΔpqsABCDE strain carrying pqsE on the pUCP18 plasmid under the Plac promoter. (C) Schematic of the RhlR-dependent QS system: RhlI makes C4-HSL (circles), and here, PqsE is discovered to be required for alternative-ligand (stars) synthesis. The two black horizontal lines represent the cytoplasmic membrane. Class I, Class II, and Class III denote RhlI-dependent, partially dependent, and independent genes, respectively. Class II and III genes require the alternative ligand for full expression. (D) Relative expression of the representative RhlR-regulated genes phzH, rhlA, and hcnA measured by qRT-PCR in the WT and designated mutants grown planktonically to HCD (OD600 = 2.0). Data are normalized to 5S RNA levels. AU denotes arbitrary units. (B and D) Error bars represent SEM for three biological replicates.