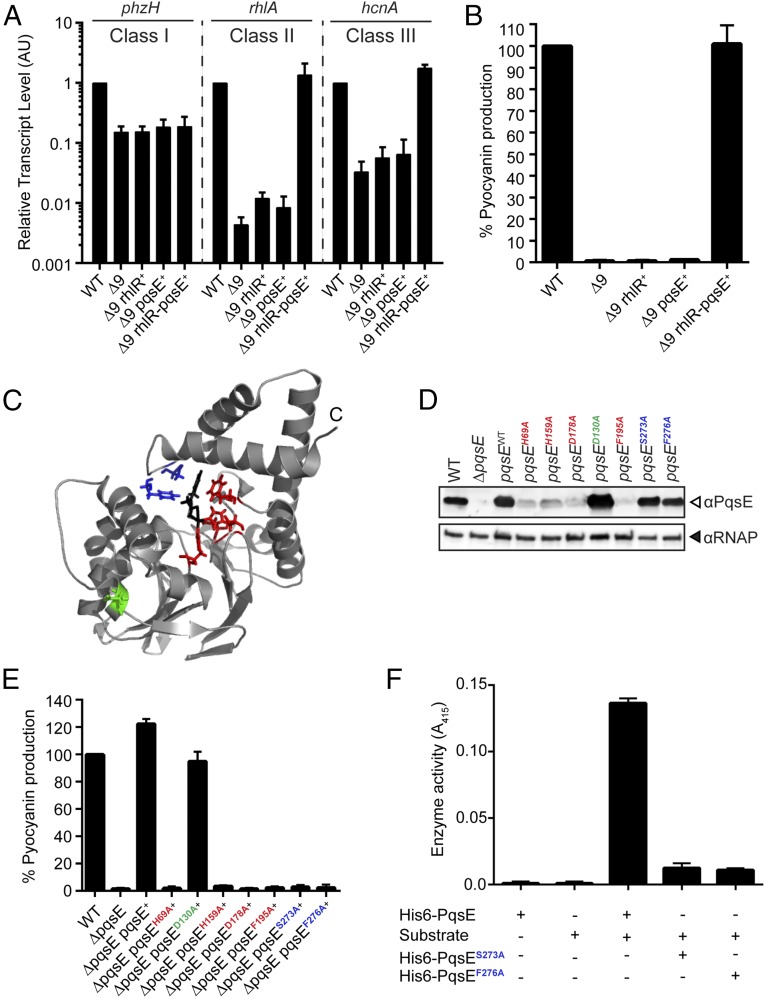
Fig. 2.
PqsE and RhlR are an autoinducer synthase–receptor pair. (A) Relative expression of the RhlR-regulated genes phzH, rhlA, and hcnA measured by qRT-PCR in the WT and designated mutants grown planktonically to HCD. Class I, Class II, and Class III are as in Fig. 1. Data are normalized to 5S RNA levels. AU denotes arbitrary units. (B) Pyocyanin production (OD695) was measured in WT PA14 and the designated mutants. (C) PqsE mutations mapped on the available structure of PqsE (gray) bound to 2-aminobenzoylacetate (black) (PDB ID: 5HIO) (24). Residues highlighted in red (H69, H159, D178, F195) form the metal ion center, S273 and F276 are required for alternative-ligand synthesis (blue), and the control residue for mutation is D130 (green). (D) Western blot analysis of lysates from WT PA14, the ΔpqsE mutant, and the ΔpqsE strain complemented with either WT pqsE or the indicated pqsE mutants. The blot was separately probed with anti-PqsE and anti-RNAP antibodies. (E) Pyocyanin production (OD695) was measured in WT PA14 and the designated mutants. (F) In vitro thioesterase activity assay for WT PqsE and the designated mutant proteins using the artificial substrate S-(4-nitrobenzoyl) mercaptoethane. In A, B, E, and F, error bars represent SEM for three independent replicates.