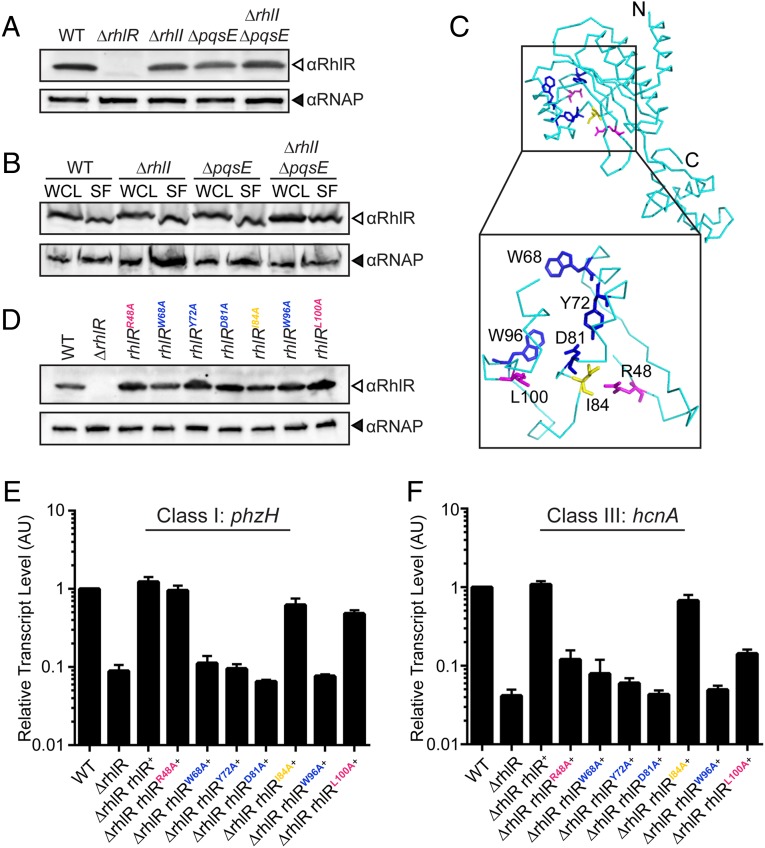
Fig. 4.
The RhlR ligand-binding domain is required for sensing the alternative ligand. (A) Western blot analysis of lysates from WT, ΔrhlR, ΔrhlI, ΔpqsE, ΔrhlI, ΔpqsE PA14 strains. RhlR levels were detected using anti-RhlR antibody, and RNAP was probed as the loading control using anti-RNAP antibody. (B) Western blot analysis of whole-cell lysates (WCL) and soluble fractions (SF) from the indicated strains; all were additionally deleted for rhlR and carry rhlR on the pUCP18 plasmid under the Plac promoter. (C) The predicted ribbon structure of the RhlR monomer (cyan) based on Phyre2 threading and comparison with the crystal structure of the closest homolog, SdiA, from E. coli (PDB ID: 4Y15) (30, 38). (Inset) Amino acids lining the putative RhlR ligand-binding pocket that were mutated in the present work. Residues in blue (W68, Y72, D81, W96) are required for response to both C4-HSL and the alternative ligand; residues in magenta (R48, L100) are required for sensing the alternative ligand but not C4-HSL, whereas reside I84 (yellow) is dispensable for detection of both ligands. (D) Western blot analysis of lysates from WT PA14, the ΔrhlR mutant, and the ΔrhlR mutant complemented with either WT rhlR or the indicated rhlR point mutants. (E) Relative expression of the RhlI-dependent phzH gene measured by qRT-PCR in the WT and mutant strains grown planktonically to HCD. Data are normalized to 5S RNA levels. Error bars represent SEM of three biological replicates. AU denotes arbitrary units. (F) As in E showing expression of the PqsE-dependent hcnA gene.