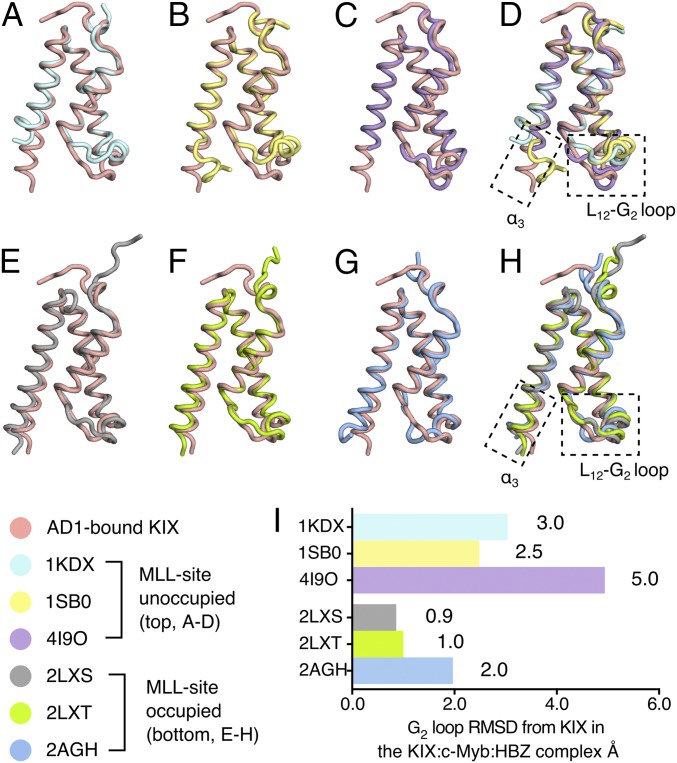
Fig. 5.
The structure of KIX in ternary complexes resembles the MLL-bound form. (A–D) Structure of AD1-bound KIX in the KIX:c-Myb:HBZ (3–77) ternary complex (pink) superimposed with KIX:pKID binary complex (PDB ID code 1KDX) (A), KIX:c-Myb binary complex (PDB ID code 1SB0) (B), and KIX (C) covalently tethered to small molecule ligand 1–10 (PDB ID code 4I9O). (D) Superposition of the AD1-bound KIX structure with KIX structures in complexes in which the MLL site is unoccupied (1KDX, 1SB0) or occupied by a small molecule ligand (4I9O) reveals large structural differences in the C-terminal region of the α3 helix and in the L12-G2 loop. All ligands are omitted for clarity. (E–H) Superposition of the structure of AD1-bound KIX in the KIX:c-Myb:HBZ (3–77) complex (pink) with the structures of KIX in the KIX:MLL binary complex (PDB ID code 2LXS) (E), the KIX:pKID:MLL ternary complex (PDB ID code 2LXT) (F), and the KIX:c-Myb:MLL ternary complex (PDB ID code 2AGH) (G). (H) Superposition of the structure of KIX bound to the HBZ AD1 domain and the structures of KIX in ternary complexes with bound MLL (2LXS, 2LXT, and 2AGH). The α3 helix of KIX is well formed and extended and the L12-G2 loop adopts a similar conformation in all four structures with the MLL site occupied. (I) Local rmsd (Å) of the L12-G2 loop regions (residues 611–620, computed using all backbone heavy atoms) between AD1-bound KIX and the structure of KIX in other complexes. In making comparisons with solution structures, the local rmsd values were calculated using the average of all structures in the ensemble.