Significance
The PI3K-Akt-mTOR pathway integrates signaling information from various mitogen and nutrient sensors to regulate cell growth and proliferation. Hyperactivation of this pathway has been observed in a number of diseases, including many cancers. Pharmacological inhibition of mechanistic target of rapamycin (mTOR) causes immediate suppression of cell-wide protein synthesis; interestingly, this effect is not uniform across all transcripts, but is biased toward mRNAs encoding ribosomal proteins. In this study, we developed a rapid, scalable, and targeted assay for RNA translation that enabled us to identify translation modulators of ribosomal proteins that act independent of mTOR signaling. These modulators include chemical compounds, acting via their “off-target” effects, as well as certain metabolic perturbations, and together, they represent a noncanonical mode of regulating ribosome biogenesis and protein synthesis capacity.
Keywords: translation profiling, translation control, mTOR, GCN2-eIF2α, ribosomal proteins
Abstract
The PI3K-Akt-mTOR signaling pathway is a master regulator of RNA translation. Pharmacological inhibition of this pathway preferentially and coordinately suppresses, in a 4EBP1/2-dependent manner, translation of mRNAs encoding ribosomal proteins. However, it is unclear whether mechanistic target of rapamycin (mTOR)-4EBP1/2 is the exclusive translation regulator of this group of genes, and furthermore, systematic searches for novel translation modulators have been immensely challenging because of difficulties in scaling existing RNA translation profiling assays. Here, we developed a rapid and highly scalable approach for gene-specific quantitation of RNA translation, termed Targeted Profiling of RNA Translation (TPRT). We applied this technique in a chemical screen for translation modulators, and identified numerous preclinical and clinical therapeutic compounds, with diverse nominal targets, that preferentially suppress translation of ribosomal proteins. Surprisingly, some of these compounds act in a manner that bypasses canonical regulation by mTOR-4EBP1/2. Instead, these compounds exert their translation effects in a manner that is dependent on GCN2-eIF2α, a central signaling axis within the integrated stress response. Furthermore, we were also able to identify metabolic perturbations that also suppress ribosomal protein translation in an mTOR-independent manner. Together, we describe a translation assay that is directly applicable to large-scale RNA translation studies, and that enabled us to identify a noncanonical, mTOR-independent mode for translation regulation of ribosomal proteins.
The PI3K-Akt-mTOR signaling pathway serves as a central regulator of cell growth and proliferation (1, 2). Intracellular signals from various mitogen and nutrient sensors congregate along this axis, and under favorable growth conditions, activation of mechanistic target of rapamycin (mTOR) kinase drives a variety of anabolic processes. Most notably, activation of mTOR, as the catalytic subunit of mechanistic target of rapamycin complex 1 (mTORC1), strongly promotes RNA translation to build up cellular protein content, and inhibition of mTOR, either pharmacologically or by withdrawal of mitogens or nutrients, leads to direct and immediate global suppression of protein synthesis. Mechanistically, mTOR inhibition leads to dephosphorylation of its substrates 4EBP1/2, permitting their association with, and sequestration of, cap-binding protein eIF4E to prevent its interaction with eIF4G at the 5′ cap of mRNAs. This ultimately leads to the well-characterized, general suppression of cap-dependent RNA translation (1, 2). Interestingly, the effects of acute, pharmacological inhibition of mTOR are not uniform among all transcripts, but instead, show a strong and preferential bias toward mRNAs that encode ribosomal proteins (RPs), all of which contain a 5′ terminal oligopyrimidine (TOP) motif, together with a small number of other transcripts that also have 5′ TOP or TOP-like motifs (3–5). To date, it remains unclear whether there exist chemical or environmental perturbations, beyond those directly targeting the mTOR pathway or its upstream factors, that are able to manipulate RP translation. The identification of such modulators could potentially provide novel avenues toward regulating ribosome biogenesis, as well as its consequential effects on protein synthesis capacity.
In this study, we asked whether there exist mTOR-independent modes of regulating RP translation. To enable a systematic search for translation modulators, we developed a rapid and scalable approach for measuring RNA translation, termed Targeted Profiling of RNA Translation (TPRT), that can interrogate a desired group of genes with very high sample throughput, and we validated this procedure using the well-characterized effects of acute mTOR inhibition and by direct comparison with ribosome profiling data. We proceeded to profile a diverse library of kinase inhibitors for their effects on RP translation, and identified a number of potential candidate compounds that suppress translation of these transcripts. On further examination, we found that two of these compounds, Dabrafenib and MK1775, modulate RP translation in a manner independent of mTOR and 4EBP1/2. We established that these compounds instead exert their translation effects via GCN2 kinase and eIF2α, which are key nodes of the eukaryotic integrated stress response. As further evidence of these mTOR-independent effects, we identified certain metabolic perturbations, namely, limitation of glucose or cysteine/cystine, that also suppress RP translation independent of mTOR activity. Together, our data point toward an alternative mode of modulating translation of RPs that bypasses canonical regulation by mTOR-4EBP1/2.
Results
An Assay for Targeted Profiling of RNA Translation.
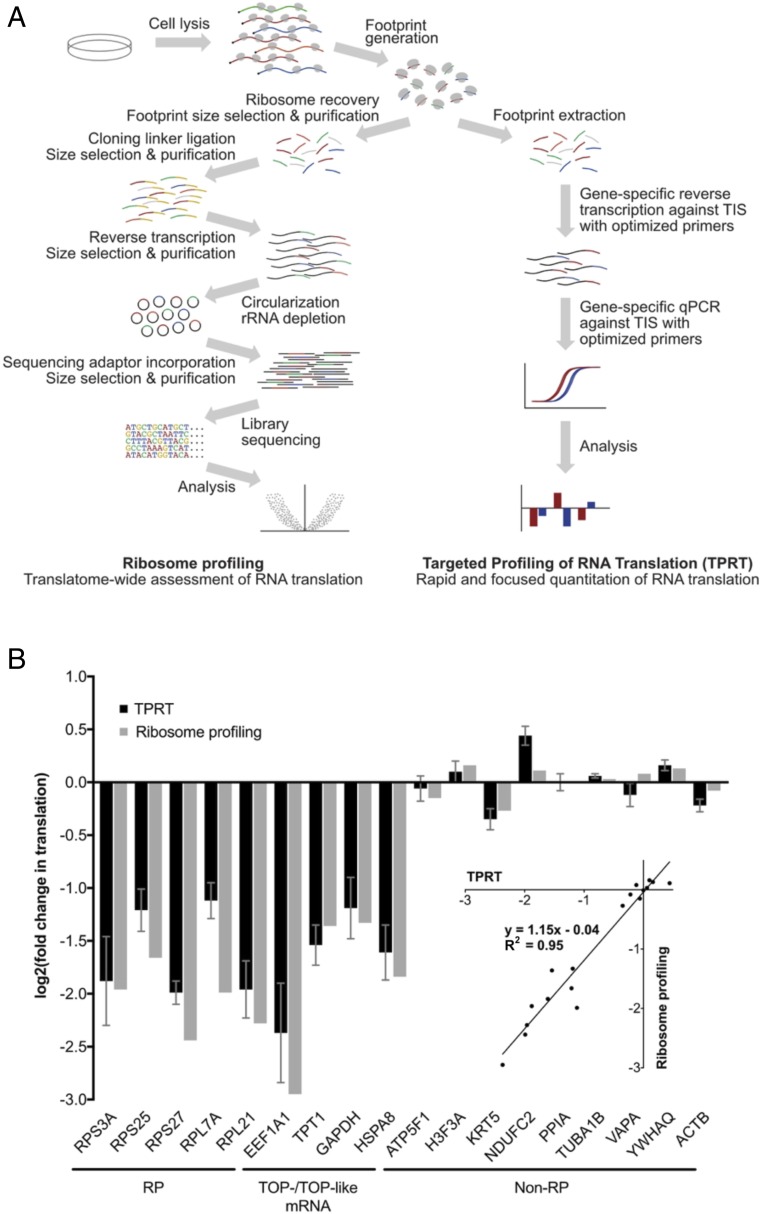
We designed and optimized a rapid and scalable RNA translation assay, TPRT, that builds on the ribosome profiling method (6, 7), and its variations for translation initiation profiling (8, 9), with the aim of simplifying sample preparation and using a gene-targeted, qPCR-based approach for translation quantitation. The TPRT procedure follows a directed workflow comprising treatment of cells with cycloheximide, rapid sample harvest, ribosome footprint extraction, reverse transcription (RT) with targeted primers against translation initiation sites (TIS) that concurrently modify cDNA template properties, and qPCR against these modified cDNA templates (Fig. 1A and SI Appendix, Fig. S1). This procedure employs a number of key optimization characteristics to improve assay behavior. First, we note that ribosome profiling examines ribosome footprint occupation distributed across the entire coding region, whereas TPRT focuses on a particular point of this region. To improve signal strength, we took advantage of the ability of certain chemical inhibitors of translation, such as cycloheximide, to artificially enrich ribosomes at TIS (6, 7, 10), and reasoned that TIS occupancy levels should be correlative with translation levels, provided sample harvest is sufficiently rapid as to prevent saturating accumulation of ribosomes at this site. We further reasoned that quantitating TIS occupation should yield greater signal strength, and therefore measurement reliability, compared with other sites. Second, given the short priming regions, the risk of unintentionally codetecting similar footprint sequences from different genes increases. We therefore employed sequence-specific primers at both the RT and qPCR stages to increase the number of selective priming steps, therefore increasing specificity against these short ribosome footprints. Third, we designed RT primers that artificially increase length of the resulting cDNA to improve detection of amplicons under fluorescence dye binding and to aid detection of primer dimer formation under melt curve analysis. In this manner, we developed a streamlined protocol for rapid and scalable quantitation of RNA translation across focused groups of genes (Fig. 1A).
Fig. 1.
Design and validation of the TPRT protocol. (A) Overview of TPRT, compared with ribosome profiling. (B) Changes in translation of RPs, other TOP- and TOP-like mRNAs, and non-RP internal controls, under Torin-1 treatment (100 nM, 1 h) in MDA-MB-468 cells, as measured by TPRT and ribosome profiling. Translation changes are relative to normalization factor of non-RP internal controls, and DMSO vehicle control, as described in SI Appendix, Materials and Methods. For TPRT, fold changes are derived as means over four biological replicates, each with four technical replicates; error bars denote SEs. For ribosomal profiling, fold changes derived as means of 2 biological replicates. (Inset) Direct comparison between TPRT and ribosome profiling mean fold changes in translation.
As validation of this assay, we applied TPRT to measure the effects of an mTOR inhibitor, Torin-1, on mRNA translation. The effects of mTOR inhibition on mRNA translation have been well characterized (3, 4), and we confirmed in MDA-MB-468 cells that acute treatment with Torin-1 at 100 nM for 1 h leads to rapid loss of 4EBP1 and 4EBP2 phosphorylation (as readouts for mTOR activity), with concomitant preferential decrease in translation of RPs and other TOP-mRNAs (SI Appendix, Fig. S2 A and B). For analysis with TPRT, we selected a panel of five RP transcripts, four non-RP transcripts that have TOP or TOP-like motifs, and nine non-RP transcripts that do not have TOP motifs (11) (SI Appendix, Table S1), and designed suitable RT and qPCR primers according to our optimized design principles (SI Appendix, Fig. S1 and Table S2). We observed that mRNA translation changes as measured by TPRT are comparable to those measured by ribosome profiling (Fig. 1B). Furthermore, given that the TPRT protocol is abbreviated to omit density- and gel-based ribosomal footprint purification steps that are present in the ribosome profiling procedure (7), we confirmed that omission of these steps in the TPRT protocol does not substantially affect the magnitude of changes measured (SI Appendix, Fig. S2C). We also analyzed a second cell line, the immortalized human mammary epithelial cell line HMEC-CT2, and likewise showed that Torin-1 decreases key markers of mTOR activity (SI Appendix, Fig. S2A), coupled with a preferential decrease in translation of RPs and other TOP-mRNAs, as measured by TPRT (SI Appendix, Fig. S2D).
A Chemical Screen Identifies translation Modulators of RPs.
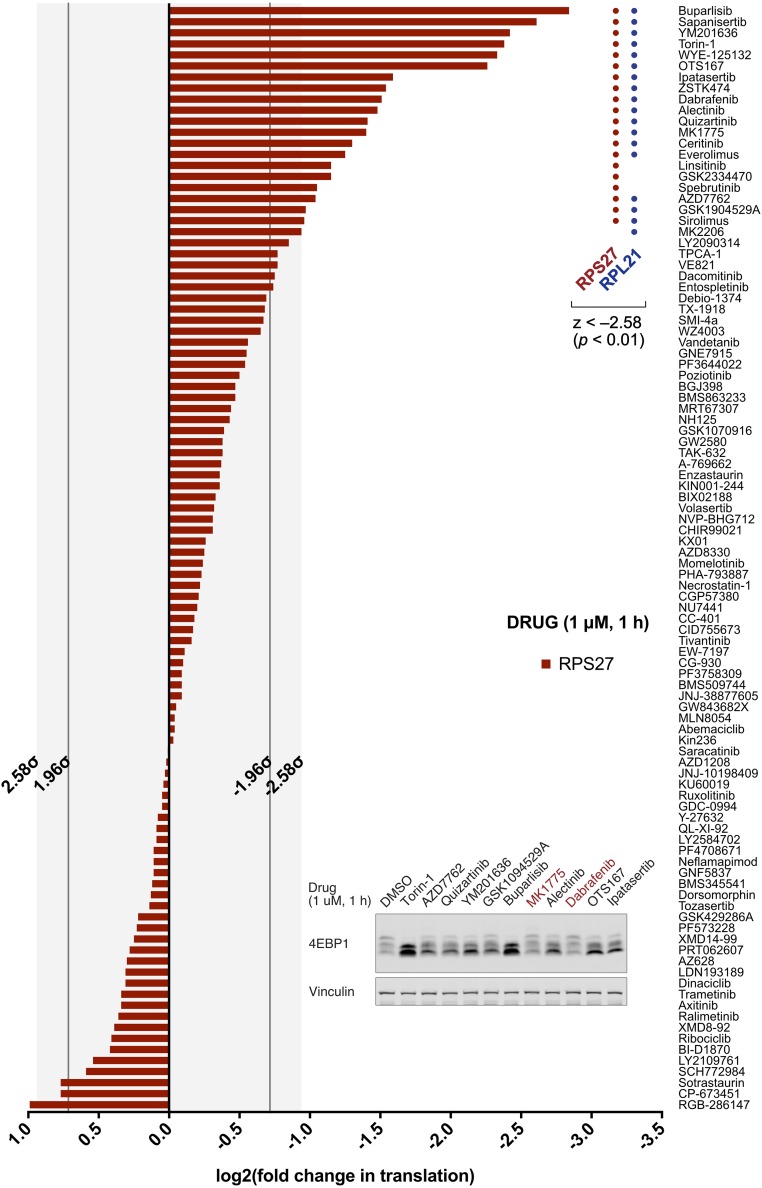
We used this assay to ask whether there are chemical modulators of RP translation beyond those targeting the PI3K-Akt-mTOR pathway. To that end, we compiled a library of 100 well-characterized and highly selective kinase inhibitors that together cover 62 distinct target families (SI Appendix, Table S3). HMEC-CT2 cells were subjected to acute treatment with each inhibitor at 1 μM for 1 h and assayed for changes in RP translation. We took advantage of the coordinated regulation of RPs by focusing on just two such transcripts for screening purposes (RPS27, RPL21), with normalization against two non-RP transcripts (PPIA, ACTB). We found that a substantial number of compounds preferentially suppress RP translation (Fig. 2 and SI Appendix, Fig. S3 and Table S3), and that many of these are preclinical and clinical compounds that have not been characterized as PI3K-Akt-mTOR inhibitors (SI Appendix, Table S4). We next tested 10 of these compounds, encompassing a broad spectrum of nominal targets, for their effects on mTOR signaling. Notably, two of these compounds, Dabrafenib and MK1775, had very little effect on 4EBP1 phosphorylation, raising the prospect that they may exert their translation effects in a manner that bypasses mTOR-mediated signaling (Fig. 2, Inset).
Fig. 2.
Chemical screen for RP translation inhibitors. Change in translation of RPS27, under acute treatment by compound library (1 μM, 1 h) in HMEC-CT2 cells. Translation changes, measured by TPRT, are relative to normalization factor of non-RP internal controls (PPIA, ACTB), and DMSO vehicle control; normalization and z-value thresholds are described in SI Appendix, Materials and Methods (SI Appendix, Fig. S3 for supporting data). (Inset) Effects of 10 RP translation inhibitors, with diverse nominal targets, on mTOR signaling as measured 4EBP1 phosphorylation shifts. Compounds that demonstrate no substantial change in 4EBP1/2 mobility shift are highlighted in red.
Dabrafenib and MK1775 Suppresses RP Translation Independent of mTOR-4EBP1/2, but Dependent upon GCN2 and eIF2α.
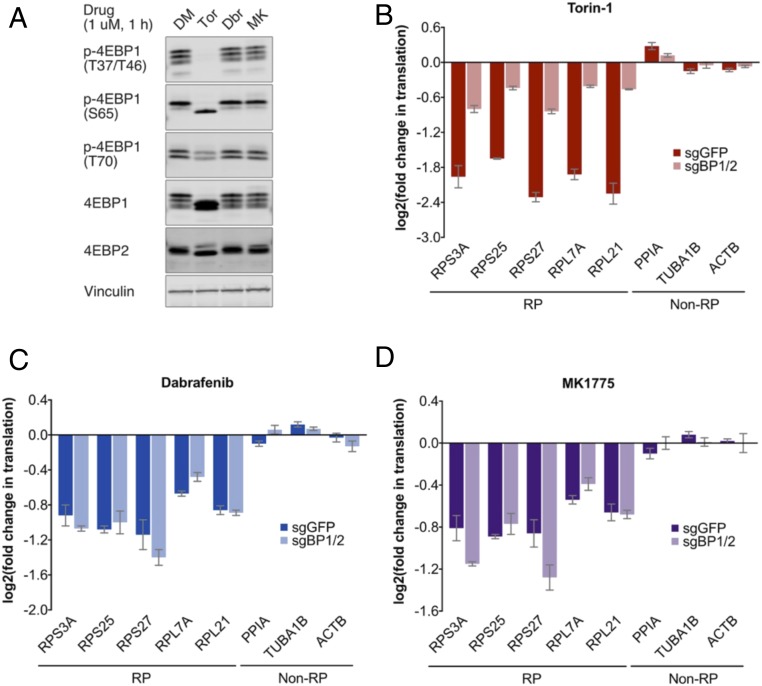
We proceeded to examine Dabrafenib and MK1775 as potential noncanonical modulators of RP translation that act independent of mTOR activity. We demonstrated that both compounds do not affect total 4EBP1 and 4EBP2 phosphorylation shifts, as determined by Western blotting, and confirmed using phospho-site-specific antibodies that phosphorylation at key functional residues is likewise unaffected (Fig. 3A). Subsequently, we used TPRT to carry out an analysis of five RP genes and three non-RP genes, and demonstrated preferential suppression of RP translation (SI Appendix, Fig. S4 A–F). Given that mTOR exerts its translation effects via 4EBP1/2 (3, 4), we asked whether Dabrafenib and MK1775 also act independent of these mTOR effectors. To that end, we generated 4EBP1/2 double-knockdown cell lines by CRISPR-Cas9 technology (SI Appendix, Fig. S4G). We found that 4EBP1/2 knockdown substantially abrogates the effects of mTOR inhibition on RP translation, as expected, but importantly, that their knockdown does not disrupt the effects of Dabrafenib and MK1775 (Fig. 3 B–D and SI Appendix, Fig. S4 H–J). Together, these observations demonstrate that both compounds suppress RP translation in an mTOR-4EBP1/2-independent manner.
Fig. 3.
Validation of Dabrafenib (Dbr) and MK1775 (MK) as mTOR-4EBP1/2-independent translation inhibitors. (A) mTOR signaling under acute drug treatment (1 μM, 1 h) in HMEC-CT2 parental cells. mTOR activity is measured by phosphorylation shifts in total 4EBP1 and 4EBP2 blots, and band intensities in phosphosite-specific p-4EBP1 blots. (B–D) Changes in RP translation under acute drug treatment (1 μM, 1 h) in HMEC-CT2 subjected to CRISPR-Cas9 targeting GFP or 4EBP1/2 (SI Appendix, Fig. S4 G–J for supporting data). Translation changes, measured by TPRT, are relative to normalization factor of non-RP internal controls, and DMSO vehicle control, as described in SI Appendix, Materials and Methods. Fold changes are derived as means over three biological replicates, each with three technical replicates; error bars denote SEs.
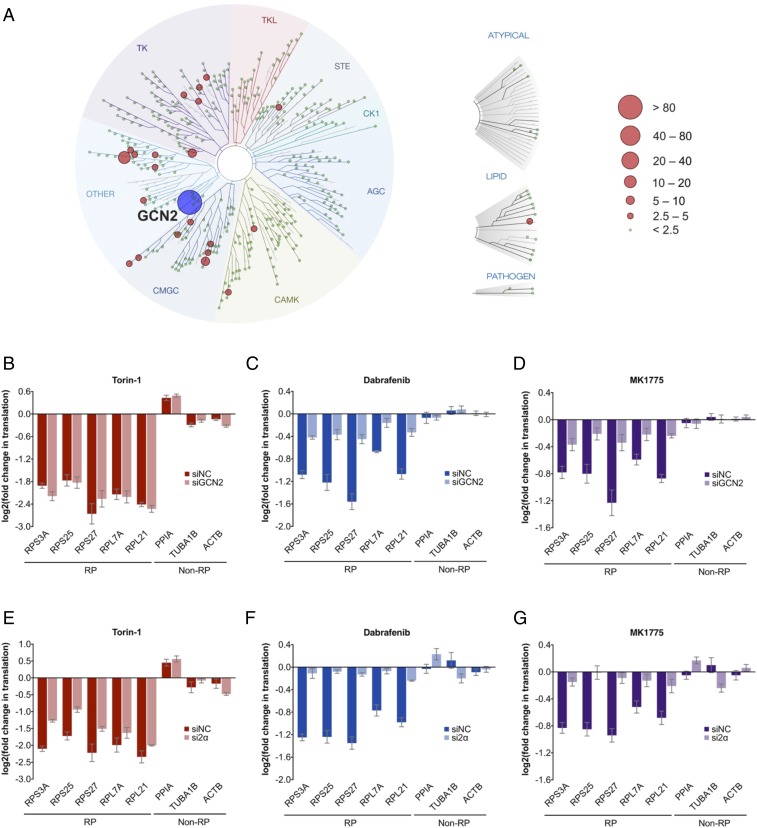
To identify the mechanistic target responsible for this effect, we first used the KINOMEscan platform to define the collection of kinases that these compounds directly modulate (12). This in vitro biochemical assay measures the extent to which a given chemical compound disrupts the interaction between a panel of recombinantly expressed kinases or kinase domains (403 wild-type kinases and 65 mutant kinases) and artificial ligands that bind to their active sites. In this manner, the KINOMEscan assay identifies the most likely kinase targets for the compound. We profiled Dabrafenib, MK1775, and AZ628, a BRAF inhibitor similar to Dabrafenib, but that does not affect RP translation (Fig. 2 and SI Appendix, Fig. S3). We reasoned that the mechanistic target may be strongly and commonly modulated by both Dabrafenib and MK1775, but remains unaffected by AZ628. We therefore computed a “likelihood score” for each kinase target, based on how closely it resembles these criteria (Fig. 4A SI Appendix, Materials and Methods and Fig. S5). Interestingly, we identified the highest-scoring candidate as GCN2, a highly conserved kinase and a central node of the integrated stress response, which regulates RNA translation in response to cellular and environmental stresses (13). Furthermore, it is well known that GCN2 serves as a kinase for activation of its downstream effector eIF2α. However, it is notable that GCN2-induced preferential RNA translation, as part of the integrated stress response, is typically restricted to genes with specialized uORF structures, such as ATF4, and has not been described as preferentially regulating RPs. To test whether the GCN2-eIF2α axis serves as the mechanistic target responsible for our observations, we subjected HMEC-CT2 cells to siRNA-mediated knockdown of GCN2 or eIF2α expression (SI Appendix, Fig. S6 A and B). We found that depletion of either GCN2 or eIF2α strongly alleviates the effects of Dabrafenib and MK1775 on RP translation (Fig. 4 B–G and SI Appendix, Fig. S6 C–H), demonstrating that the GCN2-eIF2α axis is able to preferentially regulate RP translation as an alternative mechanism that bypasses mTOR signaling. Interestingly, we found that GCN2 phosphorylation increases in a dose-dependent manner on treatment with Dabrafenib or MK1775 (albeit decreasing at very high doses), suggesting that these drugs serve to activate GCN2 kinase activity (SI Appendix, Fig. S6I). However, rather unexpectedly, it appears that phosphorylation of eIF2α, the canonical downstream target of GCN2, increases in a manner independent of GCN2 (SI Appendix, Fig. S6J).
Fig. 4.
Dependency of Dabrafenib (Dbr) and MK1775 (MK) translation effects on GCN2 and eIF2α. (A) TREEspot visualization of likelihood score (LS) for targets modulated by Dabrafenib and MK1775, and not by AZ628 (SI Appendix, Fig. S5 for target modulation by individual compounds, SI Appendix, Table S5 for LS values, and SI Appendix, Materials and Methods for LS definition). (B–G) Changes in RP translation under acute drug treatment (1 μM, 1 h) in HMEC-CT2 subjected to negative control siRNA, or siRNA targeting GCN2 (B–D) or eIF2α (E–G) (SI Appendix, Fig. S6 A–H for supporting data). Translation changes, measured by TPRT, are relative to normalization factor of non-RP internal controls, and DMSO vehicle control, as described in SI Appendix, Materials and Methods. Fold changes are derived as a mean of four (B–D) or three (E–G) biological replicates, each with three technical replicates; error bars denote SEs.
Certain Metabolic Perturbations also Suppress RP Translation in an mTOR-Independent Manner.
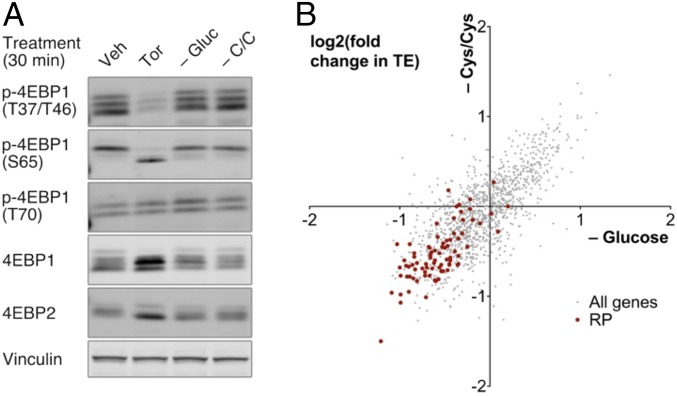
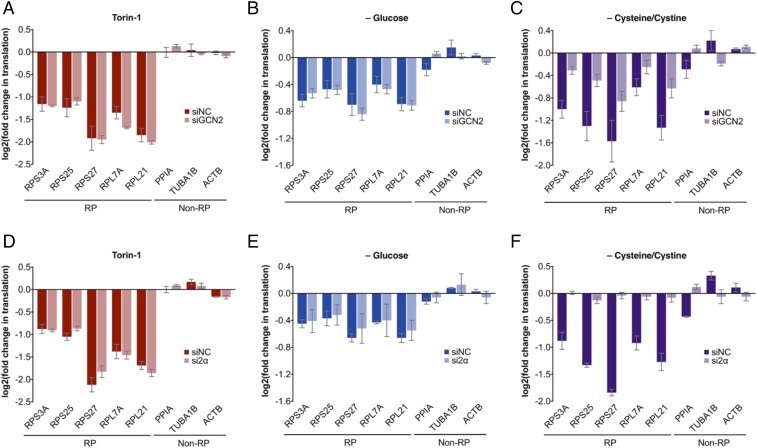
Given that mTOR and GCN2-eIF2α are both key convergence points for nutrient and stress sensors (2, 13), we asked whether there exist physiological perturbations that elicit mTOR-independent RP translation modulation. In the course of such studies, we found that in an Src-transformed MCF10A mammary epithelial model (14, 15), acute withdrawal of certain nutrients, including glucose or cysteine/cystine, causes preferential decrease in RP translation, without eliciting substantial changes in mTOR signaling (Fig. 5 and SI Appendix, Fig. S7). In line with our pharmacological findings here, we found that GCN2 or eIF2α depletion alleviates the RP translation effects of cysteine/cystine limitation, but that, interestingly, their depletion has no substantial effect on RP translation effects of glucose limitation (Fig. 6 and SI Appendix, Fig. S8 A–H). These observations demonstrate that cysteine/cystine limitation exerts its effects on RP translation through the GCN2-eIF2α axis, whereas cellular sensing of glucose deprivation occurs in a manner that is largely independent of both mTOR and GCN2-eIF2α. Furthermore, similar to the pharmacological perturbations described here, we found that cysteine/cystine limitation likewise increases GCN2 phosphorylation (SI Appendix, Fig. S8I). However, subject to some experimental variability, this increase in eIF2α phosphorylation appears to be largely independent of GCN2 (SI Appendix, Fig. S8J).
Fig. 5.
mTOR signaling and RP translation under limitation of glucose (–Gluc) or cysteine/cystine (–C/C). (A) mTOR signaling under Torin-1 treatment (1 μM, 30 min), or acute metabolic perturbations (30 min), in Src-transformed MCF10A cells. mTOR activity is measured by phosphorylation shifts in total 4EBP1 and 4EBP2 blots, and band intensities in phosphosite-specific p-4EBP1 blots. (B) Changes in transcriptome-wide translation efficiency under acute metabolic perturbations (30 min) in Src-transformed MCF10A cells. TE changes, measured by ribosome profiling, are filtered for genes with at least 100 reads per million reads in both total and ribosome footprint mRNA samples, and were computed as described in SI Appendix, Materials and Methods; RPs are highlighted in red.
Fig. 6.
Dependency of glucose (–Gluc) or cysteine/cystine (–C/C) limitation-mediated translation effects on GCN2 and eIF2α. Changes in RP translation under acute Torin-1 treatment (1 μM, 30 min), or acute metabolic perturbations (30 min) in Src-transformed MCF10A cells subjected to negative control siRNA or siRNA targeting (A–C) GCN2 or (D–F) eIF2α (SI Appendix, Fig. S8 A–H for supporting data). Translation changes, measured by TPRT, are relative to normalization factor of non-RP internal controls, and DMSO vehicle control, as described in SI Appendix, Materials and Methods. Fold changes derived as a mean of four (A–C) or three (D–F) biological replicates, each with three technical replicates; error bars denote SEs.
Together, these observations reinforce the notion of mTOR-independent means of regulating RP translation, and moreover, suggests that such regulation may be an important and prevalent mechanism by which cells respond to environmental changes.
Discussion
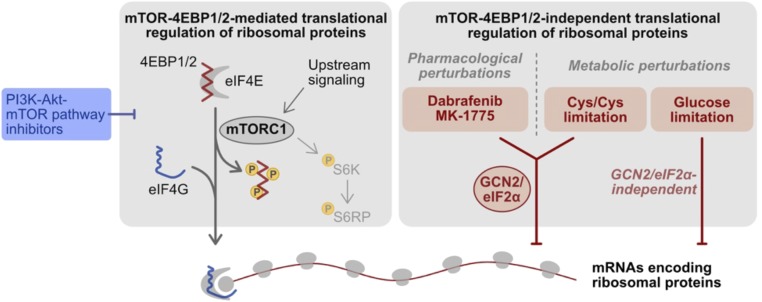
In this study, we developed a qPCR-based assay that enables time- and cost-effective interrogation of RNA translation in a gene-targeted manner, with much higher sample throughput than previously possible. We demonstrated the applicability of TPRT in a chemical profiling study to identify modulators of RP translation. Interestingly, many of these compounds have nominal targets beyond the core PI3K-Akt-mTOR pathway, raising the prospect that many more drugs than previously anticipated may exert their therapeutic effects via modulation of RNA translation. We proceeded to apply TPRT toward subsequent mechanistic studies, and established that two of these compounds, Dabrafenib (a RAF inhibitor approved for clinical use) and MK1775 (a WEE1 inhibitor currently under clinical trials for cancer indications), preferentially suppress RP translation in a manner that is independent of mTOR and 4EBP1/2, but instead act through their “off-target” effects on the GCN2-eIF2α axis (Fig. 7). Furthermore, we identified two metabolic perturbations that also elicit mTOR-independent translation suppression of RPs, and determined that one of these stresses, namely, cysteine/cystine limitation, also acts via GCN2-eIF2α (Fig. 7). The highly conserved serine-threonine protein kinase GCN2 is a key sensor in the cellular integrated stress response, and its activation promotes translation up-regulation of stress-adaptation genes such as ATF4, coupled with general suppression of RNA translation, to protect and promote survival of cells under stress conditions (13). Our findings suggest that preferential suppression of RP translation may be an uncharacterized aspect of the integrated stress response, and that artificial manipulation of GCN2 may serve as an avenue to regulating ribosome biogenesis and protein synthesis capacity.
Fig. 7.
Schematic of chemical and metabolic perturbations identified as RP translation modulators.
Our study raises a number of additional questions for further investigation. First, we identified GCN2 as a potential mechanistic target using the KINOMEscan platform. As this biochemical assay measures the extent to which a compound of interest disrupts the interaction between a kinase and a ligand for its active site, it would be expected to identify inhibitors of these kinases (12). However, both Dabrafenib and MK1775 cause dose-dependent increases in GCN2 phosphorylation (SI Appendix, Fig. S6I), an indication of its activation state (13), suggesting that these compounds serve as GCN2 activators rather than its inhibitors. Although unexpected, it is notable that some nominal BRAF inhibitors, including Dabrafenib, can paradoxically activate BRAF in cells with wild-type kinase, such as through drug-induced BRAF dimerization and/or transactivation of dimers subunits (16, 17). Second, these two pharmacological perturbations, together with cysteine/cystine limitation, modulate RP translation in an eIF2α-dependent manner and increase eIF2α phosphorylation, both consistent with integrated stress response activation. However, eIF2α phosphorylation at Ser51 nevertheless increases with GCN2 depletion under identical perturbations. We therefore infer that phosphorylation of eIF2α is not solely mediated by GCN2, and that eIF2α phosphorylation, although perhaps necessary, is certainly not sufficient to regulate RP translation. Third, our study identifies an mTOR-4EBP1/2-independent mechanism for translation regulation of RPs, but did not delve into the precise molecular elements of these transcripts that enable their regulation in such manner. RP transcripts share a number of common characteristics, most notably the 5′ TOP motif, a likely regulatory element for mTOR-mediated regulation (5).
Together, we describe an expeditious approach for interrogating RNA translation across focused panels of transcripts, and using this method, we identified mTOR-independent translation modulators of ribosomal proteins, some of which act via the GCN2-eIF2α axis.
Materials and Methods
For the targeted profiling of RNA translation (TPRT) assay, cells were treated as described in the text, washed with ice-cold PBS supplemented with 100 μg/mL cycloheximide, lysed in polysome buffer supplemented with 1 mg/mL cycloheximide, snap frozen in liquid nitrogen, thawed on ice, collected, clarified by centrifugation, and diluted to 500 ng in 100 μL lysis buffer. Samples were incubated with RNase I (Invitrogen; 1,000 U) and TurboDNase (Invitrogen; 10 U) for 1 h at 4 °C, and ribosomal footprints were extracted using TRIzol reagent (Invitrogen) and purified by isopropanol precipitation. Reverse transcription was carried out using footprint-specific primers targeting the region surrounding the translation initiation site, designed as shown in SI Appendix, Fig. S1, and using SuperScript III reverse transcriptase (Invitrogen) with a ramped temperature protocol from 40 °C to 50 °C over the course of 1 h. The RNA template was hydrolyzed by NaOH, and cDNA purified by isopropanol precipitation. qPCR was carried out using a primer pair comprising a ribosome footprint-specific forward primer targeting the translation initiation site and a common reverse primer, as shown in SI Appendix, Fig. S1, using the SYBR-Select master mix (Applied Biosystems). The TPRT method, and its variations with ribosome footprint purification and for assessing mRNA levels, is described in greater detail in SI Appendix, Materials and Methods.
Detailed information on cell lines, gene expression manipulation by CRISPR-Cas9 and siRNA, Western blotting, ribosome profiling, data analysis, and KINOMEscan profiling are described in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the J.J.Z., T.M.R., and K.S. laboratories for thoughtful discussions, Harvard Medical School Institute for Chemistry and Cell Biology-Longwood Screening Facility and Laboratory of Systems Pharmacology for provision of compounds for chemical screening, Dana Farber Cancer Institute Molecular Biology Core Facility for sequencing services, Harvard Chan Bioinformatics Core for bioinformatics consultation, Harvard Medical School Research Computing for provision of the Orchestra high-performance computing environment, Y. Wang for gift of the LentiCRISPR v2 (sgGFP) plasmid, and Q. Tang. This work was supported by NIH Grants R35 CA210057 (to J.J.Z.), R01 CA187918 (to T.M.R. and J.J.Z.), P50 CA168504 (to T.M.R. and J.J.Z.), P50 CA165962 (to T.M.R. and J.J.Z.), R01 CA107486 (to K.S.), and the Breast Cancer Research Foundation Award BCRF-17-176 (to J.J.Z.).
Footnotes
Conflict of interest statement: B.B.L. and J.J.Z. are coinventors on patent application PCT/US2017/039001. All other authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805782115/-/DCSupplemental.
References
- 1.Morita M, et al. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14:473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849:801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao X, et al. Quantitative profiling of initiating ribosomes in vivo. Nat Methods. 2015;12:147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita R, et al. Comprehensive detection of human terminal oligo-pyrimidine (TOP) genes and analysis of their characteristics. Nucleic Acids Res. 2008;36:3707–3715. doi: 10.1093/nar/gkn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 13.Pakos-Zebrucka K, et al. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch HA, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 17.Holderfield M, Nagel TE, Stuart DD. Mechanism and consequences of RAF kinase activation by small-molecule inhibitors. Br J Cancer. 2014;111:640–645. doi: 10.1038/bjc.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.