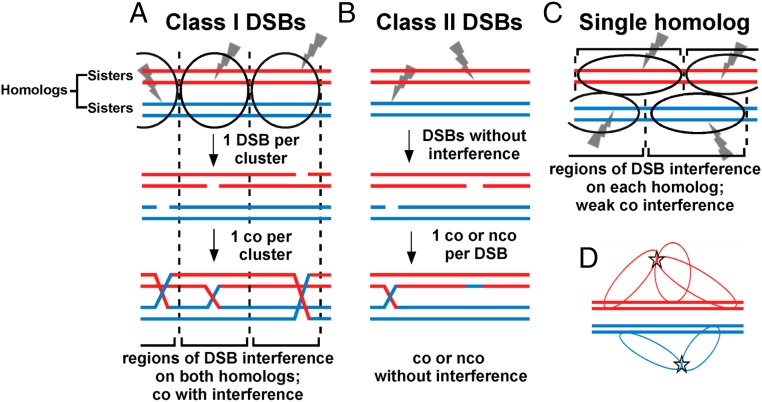
Fig. 5.
Model for crossover interference based on DSB interference among clustered DSB hotspots. Each horizontal line is one sister chromatid (dsDNA molecule), red for one parental homolog (pair of sister chromatids) and blue for the other. Ovals indicate clusters, within which one DSB (gray lightning bolt) occurs. (A) In species with complete interference, clusters of activated DSB hotspots on both homologs form in a limited chromosomal region; only one DSB (class I) is made in each cluster. Consequently, no more than one crossover is made in that region, resulting in crossover interference. In a population of cells, clusters are distributed more or less randomly; interference is thus complete in short genetic intervals but becomes less in longer genetic intervals and is negligible in genetic intervals equal to or greater than that resulting from one crossover (50 cM). (B) In some species, class II DSBs are also formed but are not cluster-controlled and consequently do not manifest interference. Crossover interference is incomplete but is greater if class I DSBs outnumber class II DSBs. (C) In some species, such as fission yeast, clusters form between activated DSB hotspot sites on one homolog, not two. Consequently, DSBs (class I) manifest interference (on one DNA molecule), but cross-overs manifest only weak interference, since DSBs form independently on the two homologs (Discussion). (D) Flexible chromatin loops enable very distant DSB hotspots bound by their activating protein determinants to form a cluster. DSB hotspots on chromatin loops could extend from the chromosomal axis (central thick lines, each a dsDNA molecule or sister chromatid) and thereby allow activated DSB hotspots to form a cluster (star), even though they might be at the extremities of the chromosome. Clusters might form over both homologs (as in A) or over only one homolog (as in C).