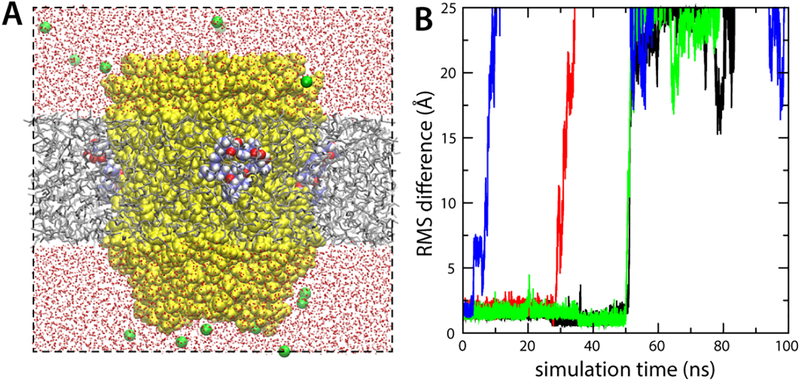
Figure 4. Oligomycin binding requires a partially aqueous environment.
(A) Simulation of the S. ceravisiae c10 ring with protonated Glu59, embedded in an artificial water-impermeable hexane layer. The protein and solvent are represented as in Fig. 2A. Initially four oligomycin molecules are bound to the cring. (B) RMS deviation of each of the four oligomycin molecules relative to their binding poses in the crystal structure, as a function of simulation time. Fast, spontaneous dissociation of all four inhibitor molecules indicates the bound state is not stable in the context of a purely hydrophobic environment.