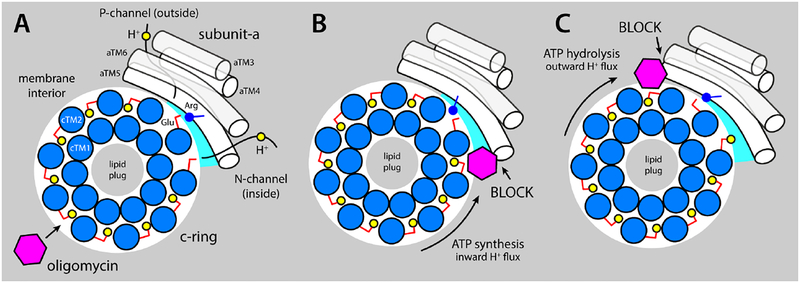
Figure 6. Proposed mechanism of inhibition of the mitochondrial ATP synthase by oligomycin.
The c-ring (marine circles) and subunit (white cylinders) are viewed from the matrix. (A) Oligomycin (magenta hexagon) binds to proton-binding sites in the c-ring that are exposed to the interior of the lipid membrane (gray area), by integrating into the coordination structure of the proton (yellow circles), which also includes the conserved Glu59 (red sticks). The c-ring rotates against subunit-a (white cylinders represent TM3-TM6, equivalent to aTM2-TM5 in bacteria), exposing two proton-binding sites to two aqueous noncollinear access permeation pathways, referred to as the P- and N-half-channels (from the positive and negative side of the membrane, respectively). A conserved arginine in aTM5 (blue stick/circle) is crucial to preclude direct proton hoping from one site to the other. Stochastic fluctuations permit one of these two sites to interact at close range with the arginine, while the other is able to load/release a proton through one of the half-channels. (B) When the c-ring rotates under a proton-motive-force, i.e. counter-clockwise, a bound oligomycin molecule eventually reaches the subunit-a/c-ring interface, blocking the release of protons into the mitochondrial matrix via the N-channel and preventing further rotation. (C) The clockwise rotation of the c-ring is halted similarly when the enzyme operates as an ATPase.