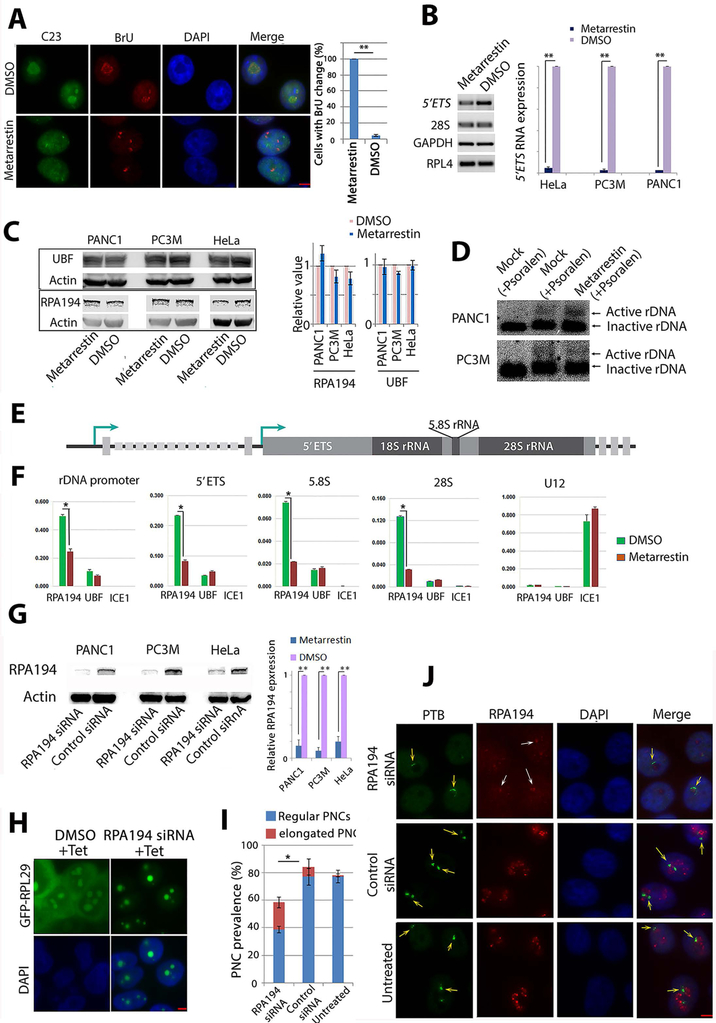
Figure 6: Metarrestin treatment reduces pre-RNA synthesis and Pol I occupancy at rDNA without changing rDNA chromatin states.
(A) Pol I transcription patterns are altered, as demonstrated by a redistribution of BrU incorporation signals from typical nucleolar labeling to pin-points at 5 min (red panel), corresponding to the changes in nucleolar structure shown as a loss of nucleolar labeling of C23/nucleolin (green panel). All cells treated with metarrestin show alterations of BrU labeling in the nucleoli compared to a small fraction of cells in DMSO treated cells (right panel). (B) RT-PCR (left panel) and qRT-PCR (right panel) show the reduction in 5’ETS of the pre-rRNA in metarrestin-treated cells. (C) Western blot analyses show no changes in protein expression of RPA194, the large subunit of Pol I, and UBF in metarrestin-treated cells. (D) Psoralen-crosslinking experiments show that the ratio of active to inactive rDNA chromatin appears unchanged upon exposure to metarrestin. (E) A diagram of rDNA structure. (F) Quantitative ChIP evaluations demonstrate that metarrestin treatment reduces the occupancy of RPA194, but not UBF, on rDNA through the promoter and the coding region. (G) Knockdown of pol I by siRNA showed reduction of RPA194 by Western blots, and the amount was quantified in relation to control siRNA treated cells (set as 1). (H) Correspondingly, ribosome synthesis was reduced in RPA194 knockdown cells, and induced GFP-RPL29 expression 72 hours after transfection of the siRNA showed the absence of cytoplasmic localization of the protein. (I) Knockdown of RPA194 by siRNA reduced PNC prevalence and increased the number of PNCs with a crescent shape (red portion). (J) RPA194 knockdown also disrupts the nucleolus (top red panel, white arrows) compared to untreated and control oligo treated cells (lower two panels). PNC structures were altered into crescent shapes (top green panel, orange arrows) compared to untreated or control oligo treated cells (lower two panels). Scale bars for all images = 5 μm. * p<0.05, ** p<0.01.