Figure 1.
Human Colonic Mesenchymal Heterogeneity in Health
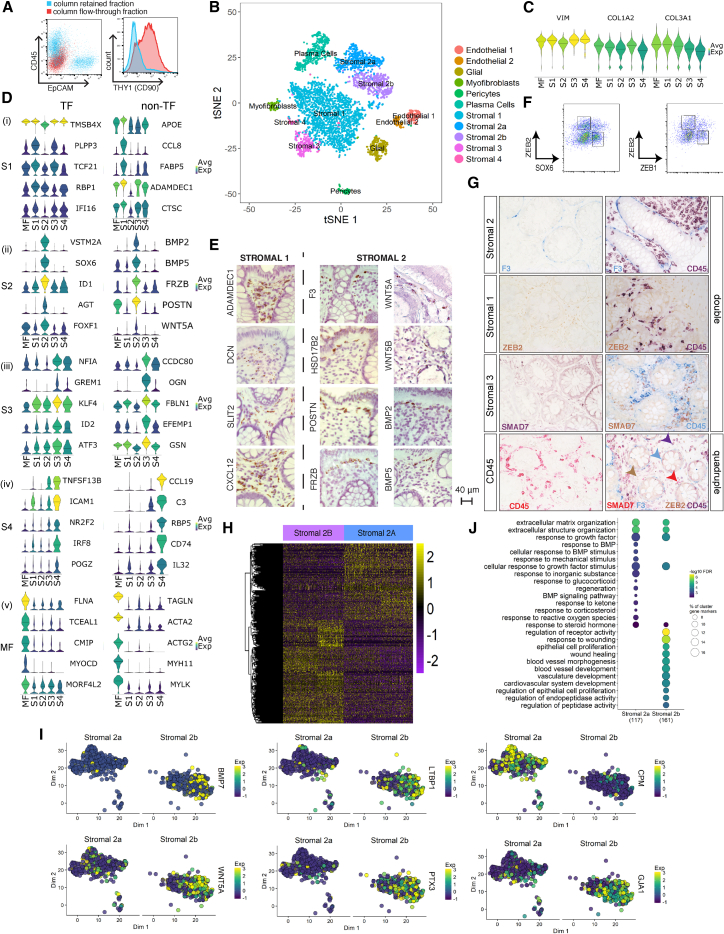
(A) Flow cytometry analysis of the indicated surface markers on colonic single-cell suspensions following removal of epithelial and hematopoietic cells by MACS. Column flow-through is shown in red, and column-retained fraction is in blue.
(B) t-SNE plot of the healthy human colonic mesenchyme dataset. Single cells colored by cluster annotation.
(C) Violin plots for pan-fibroblast marker genes vimentin (VIM) and collagen types 1 and 3 (COL1A2, COL3A1) across clusters.
(D) Violin plots for high-ranked transcriptional regulators and marker genes sharing GO annotation for significantly enriched terms for (i) S1 subset, (ii) S2 subset, (iii) S3 subset, (iv) S4 subset, and (v) myofibroblasts. Crossbars indicate median expression.
(E) Single-molecule ISH staining of healthy human colonic tissue showing distribution of S1 markers (ADAMDEC1, DCN, SLIT2, and CXCL12) (left) and S2 markers (F3 (CD142), WNT5A, HSD17B2, WNT5B, POSTN, BMP2, FRZB, BMP5) (right).
(F) Identification of SOX6−ZEB2+/ZEB1−ZEB2+ S1 and SOX6+ZEB2−/ZEB1+ZEB2− S2 subsets in healthy human colon.
(G) Single (left) and co-staining with CD45 (right) and F3/CD142 (S2), ZEB2 (S1), and SMAD7 (S3) by IHC in colonic sections. The lower far-right panel is a quadruple stain of all 4 markers.
(H) Differential expression analysis between S2a and S2b reveals 302 differentially expressed genes.
(I) t-SNE plots showing examples of genes differentially expressed between S2a and S2b.
(J) GO enrichment terms for S2a and S2b.
See also Figures S1–S3 and Tables S1–S4.