Abstract
Background
Anti–tumor necrosis factor alpha (anti-TNFα) therapy has become the mainstay of therapy for Crohn’s disease (CD). However, post-therapy, the recurrence rate is still high. The aim of this study was to dissect the molecular mechanism for recurrence of CD treated with anti-TNFα therapy and investigate novel therapeutic options that could induce complete remission.
Methods
We re-analyzed publicly available mucosal gene expression data from CD patients pre– and post–infliximab therapy to extract the transcriptional differences between responders and healthy controls. We used a systematic computational approach based on identified differences to discover novel therapies and validated this prediction through in vitro and in vivo experimentation.
Results
We identified a set of 3545 anti-TNFα therapy-untreatable genes (TUGs) that are significantly regulated in intestinal epithelial cells, which remain altered during remission. Pathway enrichment analysis of these genes clearly showed excessive growth state and suppressed terminal differentiation, whereas immune components were clearly resolved. Through in silico screening strategy, we observed that MEK inhibitors were predicted to revert expression of genes dysregulated in infliximab responders. In vitro transcriptome analysis demonstrated that selective MEK1/2 inhibitor significantly normalized reference genes from TUGs. In addition, in vitro functional study proved that MEK1/2 inhibitor facilitated intestinal epithelial differentiation. Finally, using murine colitis model, administration of MEK1/2 inhibitor significantly improved diarrhea and histological score.
Conclusions
Our data revealed the abnormalities in anti-TNFα responders’ CD colons that would be cause of recurrence of CD. Also, we provided evidence regarding MEK1/2 inhibitor as a potential treatment against CD to achieve sustainable remission.
Keywords: MEK1/2, Crohn’s disease, anti-TNFα treatment, gene signature, computational approach
INTRODUCTION
Crohn’s disease (CD) is a chronic and progressive inflammatory disease of the gastrointestinal tract.1 The management of CD has evolved dramatically over the last decade with the emergence of biological therapies. Infliximab, an anti–tumor necrosis factor alpha monoclonal antibody (anti-TNFα mAb), is effective in managing the symptoms of CD and induces remission even in patients not responding to conventional treatment including mesalamine, antibiotics, corticosteroids, and immunomodulators.2, 3 Despite these advances, still more than 50% of CD patients are estimated to undergo recurrence surgery during their lifetime due to stricturing and penetrating complications.4, 5 Furthermore, the meta-analysis study showed that the risk of relapse remained significant in CD patients when discontinuation of anti-TNFα treatment was based on clinical and endoscopic remission.6 Therefore, treatments that achieve sustainable remission in CD are needed.
Although it is largely unknown what etiology is involved in the disease, recent genetic advances have revealed that CD is presumably driven by immune and nonimmune pathophysiology.7 However, most conventional CD therapies currently target only the immune components.8 Recently, the so-called “deep remission” defined as complete mucosal healing has become the ultimate end point of the therapeutic advances for CD.9 Therefore, there is a great promise in new therapeutic approaches that integrate both immune and nonimmune pathophysiological components and potentially induce greater and sustainable deep remission as a single agent or in combination with established immunosuppressants. A previous report has demonstrated that loss of barrier function identified by confocal laser endomicroscopy would be a sensitive predictor of relapse of inflammatory bowel disease (IBD) patients in remission states.10 It has also been reported that the histology of the CD patient’s ileum in remission with anti-TNFα therapy showed complete clearance of inflammation and immune cell infiltration, whereas crypt architecture remained disturbed.11 Based on this evidence, we hypothesized that there may be some structural and functional abnormalities in patients who achieve clinical remission after anti-TNFα therapy that might predispose them to recurrence.
Integrating disease analysis into drug discovery based on molecular states is critical to finding effective therapeutics for complex diseases. At the mRNA level, gene expression signature has been used in disease characterization, the drug discovery process, or as a pharmacodynamic marker with the dose-dependent cellular response.12 Recent advances in methods for gathering and analyzing transcriptome data across many technology platforms serve as the groundwork for enabling data-driven drug discovery.13 Connectivity map (CMap) has recently been applied for the repurposing of drugs.14 The approach is to look for inverse drug-disease relationships by comparing disease molecular features with drug molecular features. Notably, Cheng et al.15 reported a systematic approach to quantitatively evaluate CMap methodology and showed that it can significantly enrich true positive drug-indication pairs using an effective matching algorithm. Thus, analyzing clinically relevant molecular signatures allowed us to identify potential therapeutic targets and understand the etiology of complex diseases.
In this study, we focused on mucosal gene signatures before and after first infliximab therapy on CD patients16 and found that a series of gene signature remained unresolved even after patients achieved clinical remission with therapy. Analysis of the gene signature provided us not only insights into nonimmune abnormalities but also a rational strategy to address this mechanism. By using a systematic computational approach, we predicted the therapeutic benefit of inhibition of the MAPK pathway. We demonstrated that both treating CD and preventing disease recurrence would be deliverable by a selective MEK1/2 inhibitor. Our results suggested that targeting MEK is a novel strategy that could induce sustainable remission via normalizing molecular alterations to baseline, which in part leads to the promotion of reconstitution of intestinal epithelium.
METHODS
Microarray Data and Differential Gene Expression Analysis
In order to reanalyze the data from Arijs et al.,16 we obtained the gene expression data set GSE16879 from GEO, the NCBI Gene Expression Omnibus, which was run on the Affymetrix platform Human Genome U133 Plus 2.0. In this study, mucosal gene expression profiles were obtained from the colonic biopsies of 19 CD patients and 24 UC patients refractory to corticosteroids and/or immunosuppression before and 4–6 weeks after their first infliximab infusion and from 6 healthy controls. The response to infliximab was defined as mucosal healing with a decrease of at least 3 points on the histological score for CD17 and as a decrease to a Mayo endoscopic subscore of 0 or 1 with a decrease to grade 0 or 1 on the histological score for UC.18, 19 Probe set expression values were estimated with the frozen RMA algorithm using R/Bioconductor, the frma package, and their presence call was calculated with the MAS5 algorithm using R/Bioconductor, the affyPLM package. Probes that were called absent in >90% samples were filtered out. Last, fewer variable probes whose interquartile ranges were in the bottom 10% of all probes were filtered out. Identification of differentially expressed genes (DEGs) was conducted based on the empirical Bayes method using R/Bioconductor, the limma package. Human crypt signature generated from GSE689420 was downloaded from the Nextbio database (http://www.nextbio.com/). For statistical test, paired t tests were performed to detect the DEGs with an adjusted P value <0.05 and absolute fold change >1.2 in the Nextbio platform. Mouse crypt signature was obtained from GSE27605 on GEO data sets,21 which was run on the Affymetrix-platform Mouse Genome 430 2.0 Array. Data processing and statistical analysis were performed applying the same methods as GSE16879.
Principal Component Analysis
Principal component analysis (PCA) was performed using the “prcomp” function of the R/CRAN stats package. To determine whether the patient groups were significantly different from the healthy group, Dunnett’s tests were conducted to examine the patient vs healthy control in PC1 using the “SimTestDiff” function of the R/CRAN SimComp package. Visualization of scatter plot and boxplot was done using the R/CRAN ggplot2 package.
Pathway Enrichment Analysis
The extracted genes were evaluated in a pathway enrichment analysis on NextBio using the Gene Ontology (GO) database.
Hierarchical Clustering and Heatmap Visualization
Clustering using cosine distance and the Ward linkage method was performed using the “Heatmap” function of the ComplexHeatmap package, which is available on R/Bioconductor. For calculation of Enrichment Scores (ES), see Supplementary Materials and Methods for a detailed description of methods.
Computational Prediction
We modified the CMap database build 0214 by integrating it with the drug signatures obtained from internal microarray transcriptome data (data source is shown as “Internal”) to compute the Enrichment Scores of gene set against drugs. See Supplementary Materials and Methods for a detailed description of methods of microarray analysis, and see Supplementary Excel File for MAS5 normalized data of the internal transcriptome data set. A list of negative permuted Enrichment Scores associated with each of the small molecules in the CMap and internal data set, as calculated by Kolmogorov-Smirnov statistics, was generated against signatures of anti-TNFα therapy–untreatable genes (TUGs) and anti-TNFα therapy–treatable genes (TTGs). For computational prediction, TUG and TTG signatures were extracted from GSE16879, per the following criteria: Significant value referred to false discovery rate (FDR)–adjusted P value <0.05 and absolute fold change >1.2, and nonsignificant value referred to FDR-adjusted P value >0.05.
Cell Culture and Reagent
Human colorectal cancer cell lines LoVo and Caco-2 were obtained from ATCC (Manassas, VA, USA). LoVo cells were grown in F-12K with 10% FBS, and Caco-2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. Both cells were maintained in a humidified incubator at 37°C and 5% CO2. Compound A was synthesized at laboratories in Takeda California as a selective and potent MEK allosteric site inhibitor (IC50 of 38 nM against purified MEK1), which was orally bioavailable and efficacious in tumor xenograft models, as previously reported.22 PD0325901 was obtained from SelleckChem (Houston, TX, USA), Gemcitabine (GEMZAR) was obtained from Eli Lilly (Indianapolis, IN, USA), and Bicalutamide (Casodex tablets) was obtained from AstraZeneca (London, UK).
Amplicon Sequencing and DEG Analysis
AmpliSeq human-transcriptome libraries were constructed and sequenced in technical triplicate using the Ion Proton platform (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. Briefly, 10 ng of total RNA was reverse-transcribed using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) followed by library generation using the Ion AmpliSeq Transcriptome Human Gene Expression Kit. Libraries were diluted to 100 pM and pooled equally, with 18 individual samples per pool. The pooled libraries were multiplexed and clonally amplified by using the Ion OneTouch 2 System, and then sequenced on Ion PI chips using an Ion Proton sequencing system. Data were first analyzed by Torrent Suite and ampliSeqRNA analysis plugin to generate count data. Read count normalization was performed using the voom method, and the extraction of DEGs was conducted based on the empirical Bayes method using R/Bioconductor, the limma package.
pERK Inhibition Assay
Total and p-ERK1/2 content was measured using an ERK1/2 (pT202/Y204 + Total) enzyme-linked immunosorbent assay kit (ab176660, Abcam, Cambridge, UK) following the manufacturer’s protocol. LoVo cells (5 × 104) were incubated in a 96-well plate treated with Compound A at indicated concentrations for 1 hour. The cells were then washed with PBS and lysed in lysis buffer. Standards for ERK1/2 and p-ERK1/2 were run simultaneously in parallel. Samples were incubated in antibody solution for 1 hour with gentle shaking at room temperature. After washing, TMB substrate was added to each well and incubated for 15 minutes. Optical density was recorded at 450 nm using a plate reader.
Transepithelial Electrical Resistance Assay
To measure the effects of Compound A on the Caco-2 monolayer barrier function, Caco-2 cells were seeded at 1 × 105 cells/mL on 24-well Transwell inserts with 0.45-µm pore size (Corning Costar, Cambridge, MA, USA). After 1 day of cell culture, the cells were treated with Compound A at indicated concentrations and cultured for 7 days. The culture medium with Compound A was changed every 2–3 days. To assess the integrity of the monolayer, transepithelial electrical resistance (TEER) was monitored by measuring the transmembrane resistance using an epithelial voltohmmeter EVOM2 (LMS). The TEER value was corrected for surface area and expressed as Ohms cm2.
Caco-2 Differentiation Assay
Cells were plated in a 96-well plate at the density of 1.6 × 104 cells/well. One day after plating, the cells were treated with compounds and allowed to grow at confluence. The entire time course was performed twice, and total RNA from cells was obtained at 1, 3, and 6 days after plating. The APOA1 and LGR5 mRNA expression levels at days 3 and 6 relative to the GAPDH mRNA level at day 1 were measured by real-time polymerase chain reaction (RT-PCR).
Quantitative RT-PCR Analysis
Total RNA was isolated from cells and purified using the RNeasy 96 kit (Qiagen, Valencia, CA, USA). Quantitative real-time PCR analysis was performed on an Applied Biosystems 7900HT Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA) using the QuantiTect Probe RT-PCR Kit (Qiagen) with TaqMan probes against the target genes (Applied Biosystems). Data were analyzed according to the 2–ΔΔCt method and normalized relative to the amount of GAPDH mRNA. The normalized abundances of target mRNAs were expressed relative to the corresponding values for cells treated with DMSO. The following TaqMan probes were used: GAPDH (4310884E), APOA1 (Hs00163641_m1), and LGR5 (Hs00969422_m1).
Preparation of Enteric Compound A Microparticles
Microparticles (MPs) were fabricated using the oil-in-oil emulsion solvent evaporation method.23, 24 See Supplementary Materials and Methods for a detailed description of methods.
Animals
Studies were performed in accordance with standards for humane care, and treatment of research animals was approved by the Institutional Animal Care and Use Committee (IACUC) of Takeda Pharmaceutical Company, Ltd (Approval No. 10797). Female BALB/c mice (7 weeks of age) and C.B-17/Icr-scid/scid mice (SCID; 7 weeks of age) were purchased from Charles River Japan and CLEA Japan, respectively. They were housed in plastic cages with free access to food and water. All animals were kept under constant temperature (23 ± 3°C) and humidity (55 ± 15%) conditions with a 12-hour light/dark cycle.
Activated T-Cell Transfer Colitis Model
Total lymphoid cells were recovered from the splenocytes of BALB/c mice by lympholyte-M (Cedarlane Lab., Ontario, Canada). Then, cell suspension was treated with HLB solution (IBL) for hemolysis. Total lymphoid cells (2 × 106 cells/mL) were cultured in RPMI-1640 (WAKO, Japan) supplemented with 10% FBS (Gibco Life Technologies), Concanavalin A (4 μg/mL), and recombinant human IL-2 (10 ng/mL, R&D systems, Minneapolis, MN, USA) for 3 days. After incubation, CD4+ T cells were isolated by MACS separation systems with CD4 (L3T4) MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Activated CD4+ T cells (2 × 105 cells) from BALB/c mice were intravenously injected into SCID mice (day 0). At day 17, the diarrhea score for stool consistency was graded on a scale of 1–4 (1, normal; 2, pasty and formed; 3, pasty and unformed; 4, diarrhea).
Therapeutic Treatment of Enteric Compound A MPs and Anti-TNFα mAb in Experimental Colitis
A total of 35 mice graded for 2 and 3 were equally divided into 5 groups as follows: vehicle control, enteric Compound A MPs at 0.3 mg/kg and at 1 mg/kg, isotype mAb (HRPN; Bio X Cell), and anti-TNFα mAb (XT3.11; Bio X Cell, West Lebanon, NH, USA). Enteric Compound A MPs were suspended in 0.5% methylcellulose (WAKO) and orally administered once a day from days 17 to 27. Isotype mAb and anti-TNFα mAb were intraperitoneally injected every 4 days from day 17 at 0.1 mg/mouse, based on preliminary studies in which the maximum efficacy of anti-TNFα treatment was observed at 0.1 mg/mouse. At day 27, the diarrhea score for stool consistency was graded on a scale of 1–4. The colon of each mouse was surgically removed and rinsed with saline, and the weight was measured at day 28.
Pharmacokinetic Studies
Pharmacokinetic studies were performed using female C57BL/6J mice and activated T-cell transfer colitis mice after oral administration of enteric Compound A MPs at a dose of 1 mg/kg. See Supplementary Materials and Methods for a detailed description of methods.
Histopathological Analysis
After the mice were killed, the colon was collected from just below the cecum to the rectal site, and the lumen was washed with PBS. The distal part of the colon was placed in 4 vol% neutral buffered formalin for 24 hours, which was replaced with PBS. All tissues were embedded in paraffin, sectioned in a cross-sectional manner, and stained with hematoxylin and eosin (H&E). Histopathological evaluation was performed independently by 2 pathologists using scoring criteria (see Supplementary Table 1, which illustrates the histopathological scoring system). The score of colitis from the distal section was calculated by combining the scores of both findings (maximum score, 8) for each animal.
In Vivo Intestinal Permeability Assay
All mice were gavaged with FITC-dextran (40-mg/100-g body weight, MW 4000; Sigma-Aldrich, St Louis, MO, USA) 4 hours before death. Whole blood was collected by cardiac puncture, and plasma was fractionated from the collected blood samples via centrifugation at 5000×g for 10 minutes at 4°C. Fluorescence intensity in plasma was analyzed using a plate reader (excitation, 485 nm; emission, 535 nm). The concentration of FITC-dextran was determined from a FITC-dextran standard curve generated by serial dilution. Permeability was calculated by linear regression of sample fluorescence.
Statistical Analysis of In Vivo Data
Values were expressed as the mean ± SEM. To evaluate the data, either the Student t test or the Wilcoxon test was performed for comparison between the 2 groups. A P value <0.05 was considered significant. For comparisons of mean values among the 3 groups, a 1-tailed Williams test or Shirley-Williams test was used, and a P value <0.025 was considered significant. All data were analyzed using the SAS System for Windows (Release 9.3, SAS Institute Inc., Cary, NC, USA).
RNA Sequencing of Mouse Colon and Differential Gene Expression Analysis
The distal part of the colon was stored in RNAlater (Qiagen) at 4°C. Total RNA was prepared using Isogen II (Nippongene, Japan) and further purified by the aid of RNeasy Mini Kit column (Qiagen) and DNaseI (Qiagen) to avoid genomic DNA contamination, according to the manufacturer’s instructions. PolyA-RNA was isolated from 2 µg of total RNA using the Dynabeads mRNA Direct Purification Kit (Thermo Fisher Scientific). The polyA-RNA was then processed for library preparation following standard procedures for Ion Proton sequencing using the Ion Total RNA-Seq Kit v2 (#4476286, Thermo Fisher Scientific). Libraries were sequenced as barcoded-pooled samples on Ion PI chips using an Ion Proton sequencing system. Normalization and DEG analysis were performed applying the same methods as amplicon sequencing.
Data Availability
The raw sequence data of our study have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). The Ampli-seq data of LoVo cell line have been deposited under accession number GSE108050, and the RNA-seq data of murine colitis model have been deposited under accession number GSE108052.
RESULTS
Mucosal Abnormal Gene Expression Remains Even After Achieving Clinical Remission With Infliximab Treatment
To compare the mucosal transcriptional profiles between IBD patients and healthy controls, we analyzed the publicly available Leuven cohort (GSE16879), in which data were obtained from control, CD, and ulcerative colitis (UC) patients at the time of pre– and post–infliximab treatment.16 A clear differentiation among the samples, depending on their disease activity, was observed when applying PCA using the given log2 microarray expression data (Fig. 1A). In the case of CD patients, 9872 gene expressions were found to be statistically different from those in healthy controls (Table 1). Importantly, transcriptome data of post-therapy samples from responders were clustered together and significantly away from the healthy controls (Fig. 1A). These results suggest that the CD intestinal mucosa in responders is molecularly different from that of healthy controls in terms of gene expression. PCA analysis showed that CD and UC are indistinguishable, suggesting the molecular similarity of inflamed CD and UC colon in spite of some phenotypic differences (see Supplementary Fig. 1, which describes PCA of CD patients, UC patients, and healthy controls).
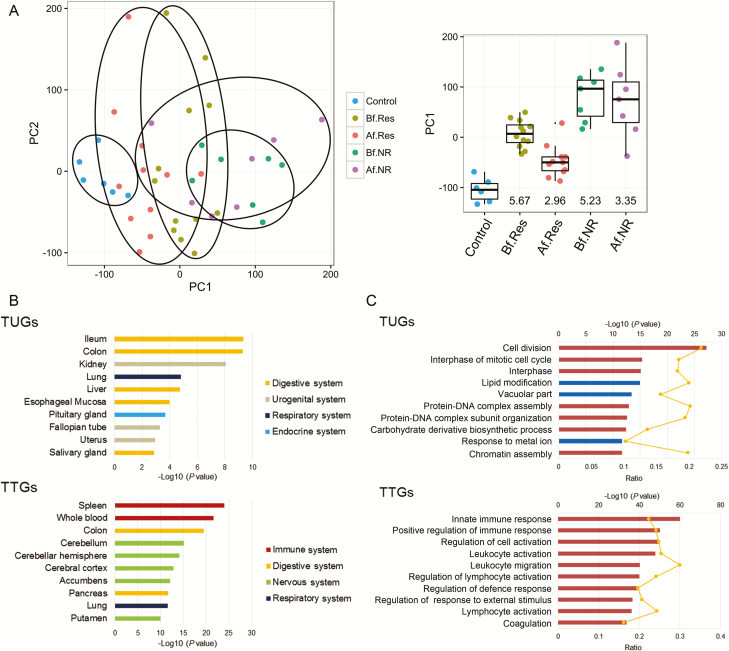
FIGURE 1.
Residual genomic profile of CD patients. A, PCA of colonic expression data from 19 patients with Crohn’s colitis (12 responders and 7 nonresponders) at the time of pre– and post–infliximab treatment and from 6 healthy controls (microarray expression data were obtained from the study by Arijs et al.16). Boxplots of PC1 distribution for each group and P values (-Log10) from the control group are as indicated on the right. B, The top 10 enriched tissue types in TUGs (upper) and TTGs (lower) using the Nextbio Body Atlas application taken from the Genotype-Tissue Expression project (GTEx) are displayed. C, The top 10 enriched GO terms in TUGs (upper) and TTGs (lower) using NextBio Pathway Enrichment application are displayed. Category names are presented on the y axis. On the x axis, the significance score (negative log of P value) for each pathway is indicated by the bars, and the line represents the ratio of genes in a given pathway that meets the cutoff criteria among total genes that make up that pathway. Red bars predict an overall increase in the activity of the pathway whereas blue bars indicate a prediction of an overall decrease in activity. Abbreviations: Af.NR, nonresponder after treatment; Af.Res, responder after treatment; Bf.NR, nonresponder before treatment; Bf.Res, responder before treatment.
Table 1:
Gene Number of Treatment Response in CD Patients
| Comparisons | Significant Genes | TUGs | TTGs |
|---|---|---|---|
| Before treatment vs healthy control | 9872 | Significant | Significant |
| After treatment vs healthy control | 5514 | Significant | Not significant |
| After treatment vs before treatment | 3602 | Not significant | Significant |
| Gene number | 3545 | 2061 | |
Significant = P value < 0.05 and absolute fold change > 1.2; not significant = P value > 0.05.
Indeed, we identified a set of 3545 genes whose expression in CD patients with remission was significantly different from healthy controls (Table 1). We designated this residual genomic expression that did not return to healthy controls as anti-TNFα therapy–untreatable genes. On the other hand, we also identified a set of 2061 genes whose expression was differently expressed from healthy controls before treatment and normalized to healthy controls in post-treatment (Table 1). We designated these recovered genes as anti-TNFα therapy–treatable genes. To examine which tissue type is potentially responsible for the TUG and TTG gene expression of CD patients, we compared the expression levels of TUGs and TTGs using the RNA sequence–based Nextbio Body Atlas application. The application allows the aggregated analysis of gene expression across >50 normal tissues of the human body system.25 We found that TUGs were highly expressed in the lower gastrointestinal tract whereas TTGs were highly expressed in immune systems such as the spleen and whole blood cells (Fig. 1B).
Next, we performed pathway enrichment analysis of TUGs and TTGs on NextBio using the Gene Ontology database. The results showed that TUGs were enriched in genes associated with cell cycle pathways whereas TTGs were enriched in inflammation and immune pathways (Fig. 1C). These results suggested that abnormal growth state in intestinal epithelial cells (IECs) persisted after achieving remission in CD colonic mucosa, whereas inflammation and immune reactions were largely resolved by infliximab therapy. To investigate whether TUGs of CD patients represent dysregulated proliferation/differentiation properties in IECs, we analyzed the association between each crypt cell gene expression signature and TUGs.20 Of interest, TUGs showed significant positive correlation with intestinal stem cells (EphB2high) vs differentiating cells’ (EphB2low) immature signature, thereby demonstrating the imbalance of self-renewal and differentiation of IECs that persists after clinical remission by anti-TNFα therapy (see Supplementary Fig. 2, a heatmap illustrating the hierarchical clustering of the human EphB2high vs EphB2low immature crypt signature compared with TUGs). These results indicate that structural and functional abnormalities may be due to unregulated self-renewal and differentiation of IECs in CD patients in remission.
In Silico Screening Identifies MAPK Pathway Inhibitors as Potential Drugs Targeting CD
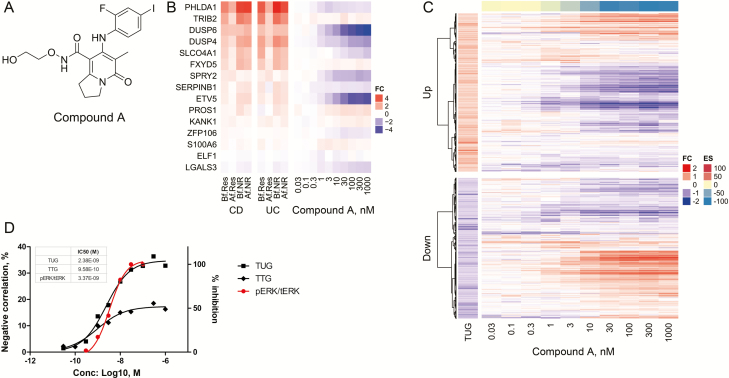
Given that TUGs remained unresolved in remission, we hypothesized that normalization of residual TUGs to healthy control could be effective in preventing recurrence of CD through correction of imbalance of IEC proliferation and differentiation. To find a novel therapeutic concept/target, we performed in silico screening to modulate the TUG signature. We used the CMap system, a systematic computational approach for drug repositioning, which is based on the integration of public gene expression signatures of drugs and diseases.26 For in silico screening, we used the modified CMap database,14 which was integrated with the drug signatures obtained from internal transcriptome data with the aim of investigating drug repositioning opportunities of internal compounds for CD treatment. For this computational prediction, we extracted TUGs and TTGs for both CD and UC by applying FDR that adjusts the P value to account for multiple hypothesis testing. This was to obtain a more robust signature and to safeguard the findings of DEGs against too many false positives creeping in. As we could identify an optimal number of TUGs for CD but not for UC by filtering with the criteria, we statistically compared extracted signatures of CD patients with each of the reference drug expression signatures from the system. As a result, we observed that MAPK pathway inhibitors (EGFR, Raf, and MEK inhibitors) were enriched and had strong negative scores with both TUGs and TTGs (Table 2). Indeed, the expressions of MEK signature genes (eg, DUSP4/6, PHLDA1, SPRY2, ETV5, etc.), which have been reported previously,27 were upregulated even in responders to infliximab treatment in CD (Fig. 2B, left). This indicates that the MAPK pathway, especially MEK, is strongly activated in CD patients, even in remission. To further investigate whether the MAPK pathway is involved in the regulation of TUGs, we used Compound A (Fig. 2A),22 a highly selective allosteric site binder of MEK1/2, among inhibitors of the Raf-MEK-ERK pathway for experimental validation.
Table 2:
Results of Gene Expression Correlation Analysis of TUGs and TTGs
| Top 20 Compounds With Negative Enrichment Score Across TUGs | |||||
|---|---|---|---|---|---|
| Rank | Data Source | Compound | Classification | Enrichment Score | Cell Source |
| 1 | Internal | Example C2555 | Raf/VEGFR inhibitor | –0.62 | COLO205 |
| 2 | Internal | Example 11456 | EGFR inhibitor | –0.56 | BT-474 |
| 3 | Internal | Compound 2cb57 | HER2/EGFR inhibitor | –0.55 | BT-474 |
| 4 | Internal | Compound 558 | Raf/VEGFR inhibitor | –0.54 | A375 |
| 5 | Internal | PD032590159 | MEK inhibitor | –0.51 | COLO205 |
| 6 | CMap | Urapidil | α1-adrenoceptor antagonist | –0.5 | PC3 |
| 7 | Internal | Gemcitabine60 | Ribonucleotide reductase inhibitor | –0.47 | Su.86.86 |
| 8 | Internal | Example 2761 | MEK inhibitor | –0.47 | A375 |
| 9 | Internal | PD0325901 | MEK inhibitor | –0.47 | A375 |
| 10 | Internal | Example 7456 | EGFR inhibitor | –0.47 | TGBC2TKB |
| 11 | Internal | Compound 3322 | MEK inhibitor | –0.46 | A375 |
| 12 | Internal | Compound 2722 | MEK inhibitor | –0.46 | A375 |
| 13 | Internal | TAK-28562 | HER2/EGFR inhibitor | –0.45 | BT-474 |
| 14 | CMap | Metergoline | 5-HT antagonist | –0.44 | PC3 |
| 15 | CMap | Flunixin | NSAID | –0.43 | PC3 |
| 16 | CMap | Alprenolol | Adrenergic beta-antagonists | –0.43 | MCF7 |
| 17 | CMap | Tiabendazole | Anthelmintic | –0.43 | MCF7 |
| 18 | Internal | TAK-73363 | MEK inhibitor | –0.43 | A375 |
| 19 | CMap | Benzamil | ENaC channel blocker | –0.42 | PC3 |
| 20 | CMap | Dicloxacillin | Antibacterial agent | –0.42 | MCF7 |
| Top 20 Compounds With Negative Enrichment Score Across TTGs | |||||
| Rank | Data Source | Compound | Classification | Enrichment Score | Cell Source |
| 1 | Internal | PD0325901 | MEK inhibitor | –0.55 | COLO205 |
| 2 | Internal | TAK-733 | MEK inhibitor | –0.4 | A375 |
| 3 | Internal | Bicalutamide64 | Androgen antagonist | –0.38 | LNCaP |
| 4 | CMap | Carbenoxolone | 11β-hydroxysteroid dehydrogenase inhibitor | –0.34 | PC3 |
| 5 | CMap | Sitosterol | Inhibit cholesterol’s absorption | –0.33 | MCF7 |
| 6 | CMap | Betahistine | H1 receptor agonist | –0.33 | PC3 |
| 7 | CMap | Iohexol | Contrast agents | –0.33 | MCF7 |
| 8 | CMap | Cinchonine | Antimalarial drug | –0.32 | PC3 |
| 9 | CMap | Dicloxacillin | Penicillin-like antibiotic | –0.32 | MCF7 |
| 10 | CMap | Methylergometrine | Oxytocics | –0.31 | PC3 |
| 11 | CMap | Co-dergocrine mesilate | Antimigraine | –0.31 | PC3 |
| 12 | CMap | Prestwick-1083 | — | –0.31 | PC3 |
| 13 | CMap | Nadolol | Adrenergic beta antagonist | –0.3 | MCF7 |
| 14 | CMap | Sulmazole | A1 adenosine receptor antagonist | –0.3 | PC3 |
| 15 | CMap | Triamterene | Sodium channel blocker | –0.29 | MCF7 |
| 16 | CMap | Ajmaline | Sodium channel blocker | –0.29 | MCF7 |
| 17 | CMap | Furosemide | Sodium potassium chloride symporter inhibitor | –0.29 | PC3 |
| 18 | CMap | Midodrine | Adrenergic alpha agonist | –0.29 | PC3 |
| 19 | CMap | 4-hydroxyphenazone | — | –0.29 | MCF7 |
| 20 | CMap | Amiprilose | Anti-inflammatory agent | –0.29 | MCF7 |
FIGURE 2.
MEK1/2 inhibitor normalized reference CD signature in LoVo cells. A, Chemical structure of Compound A. B, Heatmap showing the genes from the MEK activation signature for both CD and UC patients (left) compared with fold change in gene expression levels in LoVo treated with Compound A across 10 dose response conditions (0.03–1000 nM, n = 3, right). C, Heatmap of TUGs clustered into upregulated and downregulated genes compared with fold change in gene expression levels in LoVo treated with Compound A across 10 dose response conditions (0.03–1000 nM, n = 3), obtained by Ampli-seq analysis. The color-coded scale for the normalized expression value is indicated at the right of the figure, which correlates with color intensity to the fold change of gene expression and enrichment score. D, IC50 values were calculated by plotting both negative correlation rate between Compound A DEGs of TUGs/TTGs and in vitro pERK inhibitory activity in LoVo cells using a nonlinear regression analysis by GraphPad Prism (GraphPad, La Jolla, CA, USA).
MEK1/2 Inhibitor Normalizes Reference CD Signature In Vitro
To examine whether Compound A modulates TUG and TTG expression in vitro, we used Compound A–treated LoVo, a human colorectal cancer cell line, to compare the transcriptomes with DMSO control. We first looked at the expression of MEK signature genes and confirmed that most of these genes were downregulated in response to Compound A treatment (Fig. 2B, right). Importantly, TUGs showed a significant negative correlation with DEGs between Compound A–treated and DMSO-treated cells in a dose-dependent manner (Fig. 2C and D). This reversible effect against TUGs was almost equivalent to the value for the pharmacodynamic inhibitory effect against ERK1/2 phosphorylation in LoVo cells (Fig. 2D). On the other hand, the negative correlation rate between TTGs and the DEGs was lower than that of TUGs (Fig. 2D). Moreover, a significant negative correlation was observed between the immature signature and the DEGs in a dose-dependent manner whereas TUGs were significantly positively correlated (see Supplementary Fig. 2, a heatmap illustrating the hierarchical clustering of the human EphB2high vs EphB2low immature crypt signature compared with the Compound A–treated signature). Our important finding from this transcriptome analysis is that the MEK inhibitor significantly induced an opposite effect on the TUG signature, and the result also provided an important insight into the effect of MEK inhibitors on the pathophysiological function of IECs.
MEK1/2 Inhibitor Promotes In Vitro Caco-2 Cell Differentiation and Barrier Function
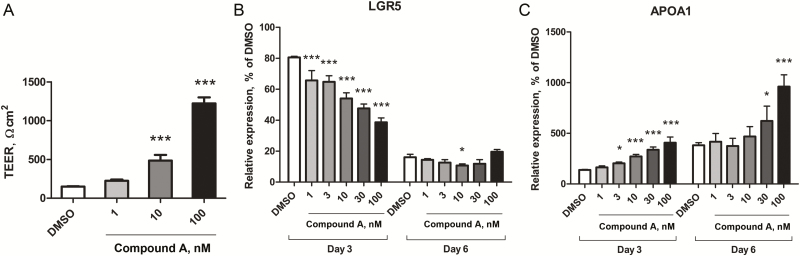
Next, we investigated whether TUG modulation by Compound A has a significant effect on the pathophysiological function of intestinal permeability resembling enterocytes. Caco-2, a human colorectal cancer cell line, was reported to acquire “matureness,” such as tight junction assembly and cell-cell adhesion–initiated polarization during 21 days in culture, and the regulation of transcription underlying Caco-2 cell polarization is similar to that of in vivo enterocyte differentiation.28 First, we evaluated the effect of Compound A on transepithelial electrical resistance (TEER) assay, which is commonly used to assess the barrier function of IECs. As shown in Fig. 3A, Compound A exhibits a significant increase in TEER over a Caco-2 monolayer in a dose-dependent manner. Next, we examined the expression of apolipoprotein A-1 (APOA1) and leucine-rich repeat–containing G-protein-coupled receptor 5 (LGR5), which are enterocyte and stem cell markers, respectively. Consistent with the results in TEER measurement, Compound A treatment increased the expression of APOA1 and decreased the expression of LGR5 (Fig. 3B and C). These results indicate that inhibition of MEK enhances IEC differentiation and maturation and epithelial barrier function.
FIGURE 3.
MEK1/2 inhibitor enhanced intestinal barrier formation by promoting differentiation of IECs. A, TEER was measured to assay the integrity of the epithelium layer formed by differentiated Caco-2 cells. The values represent the means of n = 4 samples ± standard deviations. Levels of statistical significance compared with DMSO control: ***P value < 0.0005 by Williams test. Relative mRNA expression levels of APOA1 (B) and LGR5 (C) at day 3 and day 6 were measured by RT-PCR, normalized with the expression of GAPDH at day 1. The values represent the means of n = 3 samples ± standard deviations. Levels of statistical significance compared with DMSO control: *P value < 0.025: **P value < 0.005: ***P value < 0.0005 by Williams test.
MEK1/2 Inhibitor Shows Significant Therapeutic Effect on Histology and Diarrhea in Activated T-Cell Transfer Colitis Model
To determine whether our in silico target prediction model would be translated into therapeutic efficacy in vivo, we performed an efficacy study using Compound A by means of an activated T-cell transfer colitis model.29 The adoptive transfer of activated CD4+ T cells to SCID mice promoted diarrhea within 2–3 weeks, and histological changes correlated well with the disease parameters of colitis, such as colon weight gain, stool consistency score, and the increase in colonic inflammatory cytokines. Although highly selective and potent oral MEK inhibitors have been developed and assessed in numerous clinical studies for cancer treatment, a number of mechanism-based toxicities have emerged.30 In particular, acneiform dermatitis, a serious skin rash, is a frequent side effect of MEK inhibitors including Compound A. Thus, to minimize systemic exposure leading to systemic toxicities, we developed a colon-specific drug delivery system for Compound A by conducting several formulation approaches. This also aims to enhance local therapeutic efficacy.31 Importantly, Compound A has superior characteristics for a colon-targeted formulation such as higher solubility and lower permeability, which can limit systemic absorption to prevent drug distribution into normal tissues despite its long-term retention in the colon, compared with other MEK inhibitors (eg, trametinib, selumetinib, and binimetinib). Based on a preliminary pharmacokinetic study using C57BL/6J normal mice, the area under the plasma concentration time curve (AUC) of Compound A MPs decreased by 97.32% as compared with the suspensions, whereas the concentrations in the colon of both groups were almost the same level. This demonstrated that the amount of Compound A absorbed into plasma was obviously decreased when encapsulated in the enteric MPs, thus reducing the side effects caused by systemic absorption. Moreover, in vivo colonic pharmacodynamics study using enteric Compound A MPs in LPS-treated mice demonstrated that the level of colon AUC0–24h (1206.6 ng・h/g) showed target engagement defined as pERK inhibition at the dose level whereas plasma AUC0–24h (7.2 ng・h/mL) was within safe range. Likewise, in colitis mice, Compound A MPs showed safe systemic exposure at the dose levels where therapeutic effect was observed despite repeated dosing. Further, as the colon AUC level was also almost the same as that of LPS-treated normal mice, which was enough to inhibit MEK enzymatic function, we concluded that enteric Compound A MPs were efficiently delivered to the colon.
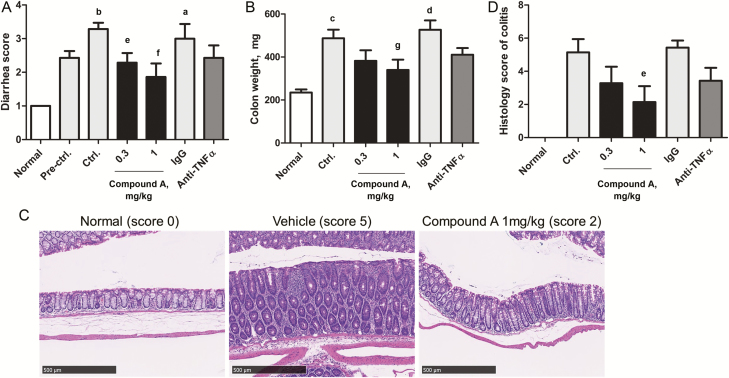
When enteric Compound A MPs were orally administrated therapeutically at the dose of 0.3 and 1 mg/kg, q.d., diarrhea score and the increase in colon weight were significantly inhibited compared with vehicle treatment (Fig. 4A and B). On the other hand, only a partial inhibition of diarrhea score and colon weight gain was observed in the mice treated with anti-TNFα mAb at the dose of 0.1 mg/mouse compared with isotype control treatment (Fig. 4A and B). Importantly, enteric Compound A MPs substantially ameliorated extensive injury of the colon mucosal layer, which was observed in the vehicle mice with histopathological analysis (Fig. 4C). Quantitative evaluation demonstrated that the histology score of colitis was significantly reduced with mucosal regeneration (Fig. 4D). Also, immune cell infiltration was significantly reduced in mice treated with Compound A at the dose of 1 mg/kg compared with the vehicle control group (see Supplementary Table 2, which illustrates the histopathological findings of the colon treated with Compound A and anti-TNFα mAb). On the other hand, the histology score of colitis was not significantly reduced in the mice treated with anti-TNFα mAb compared with isotype control treatment. We also performed in vivo permeability assay using a fluorescein isothiocyanate (FITC)–labeled dextran method. This experiment revealed that activated T-cell transfer colitis mice receiving oral treatment with Compound A tended to decrease intestinal permeability compared with untreated colitis mice (0.3 mg/kg and 1 mg/kg vs vehicle control; 72.2% and 75.0%, P = 0.055, 0.049 by 1-tailed Shirley-Williams test, respectively), whereas treatment of anti-TNFα mAb had no effect on it (vs isotype control; 133.0%, not significant). Together, these data provided in vivo evidence that enteric Compound A MPs exhibit therapeutic efficacy and improve the histological changes in the colitis model.
FIGURE 4.
Therapeutic efficacy of enteric MPs of MEK1/2 inhibitor in activated T-cell transfer colitis model. A, Diarrhea score data and (B) colon weight data were analyzed in a blind fashion. C, Representative H&E-stained colon sections of normal mice, vehicle, and 1 mg/kg Compound A treated activated T-cell transfer colitis mice (magnification, ×200, scale bar = 500 μm). D, Quantitative evaluation of the histology score of colitis was analyzed. Data are presented as the mean ± SE of 4 normal or 7 transferred mice. aP < 0.01, bP < 0.001 (Aspin-Welch test); cP < 0.01, dP < 0.001 (Student t test) vs normal group. eP < 0.025, fP < 0.005 (1-tailed Shirley-Williams test); gP value < 0.025 (1-tailed Williams test) vs vehicle control group.
MEK1/2 Inhibitor Treatment in Colitis Mice Downregulates Gene Expression That Associates With Both Dysregulated Growth in IECs and Pro-inflammatory Response in Immune Cells
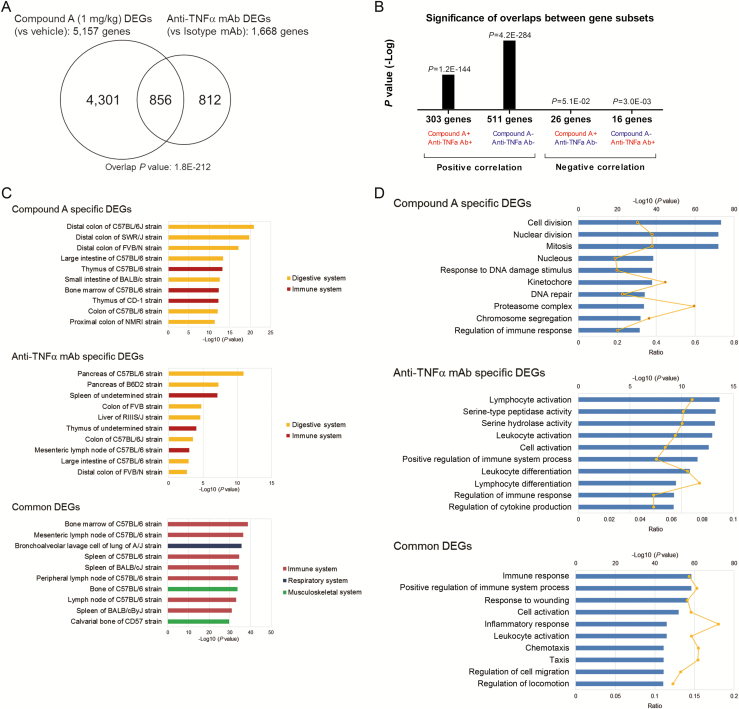
To compare transcriptome differences between Compound A–treated and anti-TNFα mAb–treated colitis mice, we examined the global gene expression profile by RNA sequencing. The Venn diagram in Figure 5A summarizes the overlapping transcripts detected in each sample. A total of 5157 genes were identified as DEGs of Compound A, which was subsequently compared with the colitis signature (vehicle vs normal mice). Of interest, 46.1% of Compound A–regulated genes were negatively correlated with colitis signature, whereas only 11.1% of anti-TNFα mAb DEGs (1668 genes) were negatively correlated with colitis signature, which reflects the superior efficacy of Compound A to anti-TNFα mAb (see Supplementary Fig. 4, a heatmap illustrating hierarchical clustering of colitis signature compared with Compound A– and anti-TNFα mAb–treated signature). We hypothesized that dynamic molecular and signaling changes might occur specifically in an activated T-cell transfer colitis model treated with Compound A. To obtain new insights, we conducted a tissue type enrichment analysis using the mouse Body Atlas database of Compound A–specific, anti-TNFα mAb–specific, and common DEGs between Compound A and anti-TNFα mAb. We found that nearly four-fifths of Compound A DEGs, which were different from anti-TNFα mAb DEGs, were significantly enriched in the lower gastrointestinal tract. In contrast, immune systems such as bone marrow, mesenteric lymph node, and spleen were enriched in common DEGs (Fig. 5C). Pathway enrichment analysis was also performed for each DEG. The results demonstrated that genes associated with cell cycle and DNA replication were significantly downregulated in Compound A, specifically treatable genes (Fig. 5D). On the other hand, common DEGs between Compound A and anti-TNFα mAb showed a significantly positive correlation (Fig. 5B). The pathway analysis of common signature revealed that inflammation and immune pathways were significantly downregulated (Fig. 5D). These results indicate that Compound A can attenuate abnormal growth in colonic mucosa by targeting IECs and exhibit an anti-inflammatory effect by targeting immune cells. Moreover, compared with mouse crypt gene expression, colitis signature was significantly positively correlated with mouse immature signature, which was extracted from publicly available mouse EphB2high vs EphB2low transcriptome data21 (see Supplementary Fig. 2, a heatmap illustrating hierarchical clustering of mouse EphB2high vs EphB2low immature crypt signature compared with Compound A–treated signature). Interestingly, the immature signature was significantly negatively correlated with DEGs of Compound A (ES, –163) whereas negative correlation with anti-TNFα mAb DEGs was not so significant (ES, –5.4). These results suggest that Compound A could repair dysregulated IECs more effectively than anti-TNFα mAb. These findings corroborated the expected mechanism of efficacy of Compound A in the colitis model, which was computationally predicted by using a gene signature derived from CD patients.
FIGURE 5.
Therapeutic efficacy of MEK1/2 inhibitor against colonic gene expressions in activated T-cell transfer colitis model. A, Venn diagrams illustrate the numbers of significantly affected genes that were shared or were uniquely changed by 1-mg/kg Compound A or anti-TNFα mAb treatment (absolute fold change > 1.2, P < 0.05). B, Almost all genes affected by both Compound A and anti-TNFα mAb were significantly positively correlated. The x axis represents the class of each overlapping correlated gene; “+” and “−” symbols depict upregulated and downregulated genes, respectively, and the y axis provides P value (-Log). C, The top 10 enriched tissue types in Compound A specific (upper), anti-TNFα mAb–specific (middle), and common DEGs between Compound A and anti-TNFα mAb (lower) using mouse the Body Atlas database. D, The top 10 enriched GO terms in each Compound A–specific (upper), anti-TNFα mAb–specific (middle), and common DEG (lower) using the NextBio pathway enrichment application. Category names are presented on the y axis. The x axis indicates the -Log10 (P value) of the over-representation analysis. Category names are presented on the y axis. On the x axis, the significance score (negative log of P value) for each pathway is indicated by the bars, and the line represents the ratio of genes in a given pathway that meet the cutoff criteria among total genes that make up that pathway. All blue bars predict an overall decrease in the activity of the pathway.
DISCUSSION
In the current study, we present evidence that may help to resolve the relapse of CD and obtain sustainable remission. CD has periods of relapse and remission, but there is no long-term cure. Surgeries are still required in some patients. For CD patients in remission, relapse rates at 1, 2, 5, and 10 years are estimated at 20%, 40%, 67%, and 76%, respectively.32 As for infliximab, Chauvin et al. reported a recurrence rate of 68% among CD patients in treatment in a 12-year retrospective study.33 This evidence suggests that there are unmet medical needs in the management of CD. For assessing the possible mechanism of relapse, an unresolved issue is whether colonic lesions of CD patients in remission continue to express abnormal transcripts or not as compared with healthy controls. We present a method to dissect the physiological response with anti-TNFα therapy by exploiting several bioinformatics analyses with a CD patient–relevant mucosal gene signature. We have defined a unique gene signature “TUGs,” which did not return to a “healthy” state even after achieving clinical remission. Importantly, TUGs are positively correlated with the immature signature of IECs, suggesting that the residual transcripts are involved in expansion of undifferentiated cells and decrease or loss of mature functional IECs in infliximab responders (see Supplementary Fig. 2, a heatmap illustrating hierarchical clustering of the human EphB2high vs EphB2low immature crypt signature compared with TUGs). Recently, it has been proposed that dysregulation within a differentiation system for correct IEC formation perpetuates impaired epithelial homeostasis and plays a crucial role in IBD pathogenesis by increasing intestinal permeability.34–37 Furthermore, in the case of CD patients, an increase in intestinal permeability has been reported to precede episodes of disease relapse and the onset of symptoms by up to 1 year.38, 39 However, it is not yet clear what molecular events are responsible for dysregulated homeostasis of IECs that predispose to relapse in patients with quiescent CD. Considering that perturbations of IECs homeostasis can lead to intestinal disorders,40 we hypothesized that TUGs might lead to increased intestinal permeability and distorted intestinal mucosal barrier function, ultimately causing recurrence. Similar strategies to extract unresolved gene signature have been previously tried in patients with psoriasis41 and UC,42 but the gene signature associated with CD remission was not studied.
In this study, we inferred that the MEK inhibitor would be a potential new therapeutic agent for CD based on the results of an in silico computational approach using TUGs. Previous reports have demonstrated that ERK1/2 activation may have importance for diarrhea in CD patients.43 It has also been reported that constitutive activation of the MEK/ERK cascade inhibits enterocyte differentiation, in part through inhibition of the transcriptional activity of CDX2, a master transcription factor regulating the differentiation, cell-cell adhesion, and polarity of IECs.44–46 Conditional homozygotes in Cdx2 knockout mice have been previously shown to exhibit loss of intestinal morphology and cause a serious disruption in the mucosal architecture.47 Interestingly, we observed that transcripts linked to intestinal differentiation including several CDX2 target genes44 were upregulated following MEK inhibition in vitro (see Supplementary Fig. 3, a heatmap showing the genes from the CDX2 target genes for both CD and UC patients and compared with the Compound A–treated signature). This finding is consistent with the role of CDX2 as a master regulator of intestinal morphogenesis and suggests that genes controlling epithelial differentiation are suppressed by constitutive activation of the MEK/ERK pathway. Although we cannot conclude what the physiological consequences of reversing TUGs to a normal state are, it could be speculated that MEK inhibitor partly enhances CDX2 transcriptional activity, which in turn enhances epithelial differentiation and finally leads to restitution of dysregulated IECs.
Indeed, MEK inhibitor exhibited a significant increase of the TEER using the Caco-2 cell line, which is a widely used intestinal cellular model that retains very similar morphologic properties to enterocytes (Fig. 3A). However, using transformed cell lines has some limitations.48 A major limitation is that intestinal cells growing in 2D monoculture lack the physiological extracellular matrix microenvironment that is necessary to maintain in situ phenotypes. To overcome this limitation, the utility of in vitro 3D organoids that effectively mimic IEC structure and functional features has been well confirmed for testing epithelial differentiation with the capacity to model heterogeneous cell fates.49 Therefore, further studies will be necessary to clarify the effect of MEK inhibitor on intestinal differentiation using an organoid model for drug validation. Nevertheless, although animal models also incompletely recapitulate the complex pathophysiology of the intestinal diseases of humans, the histology score was significantly reduced in response to MEK inhibitor treatment in the in vivo colitis model in this study (Fig. 4D). These results provide important insight into the beneficial effect of MEK inhibitor on promoting intestinal barrier function.
In addition, interestingly, in silico computational screening revealed that several MEK inhibitors scored high in not only the TUG signature but also the TTG signature. From this result, MEK inhibitor would have a potential therapeutic effect to reverse both TNFα-untreatable and -treatable gene expression. MEK inhibitor would have great promise in new therapeutic approaches that integrate both immune and nonimmune pathophysiological components and potentially induce greater and sustainable remission as a single agent. Indeed, transcriptome analysis showed that Compound A reverses a set of genes that are not only cell cycle–related genes, which are highly expressed in IECs, but also inflammation-related genes, which overlapped with DEGs of the anti-TNFα mAb in vivo model (Fig. 5C). Moreover, a previous report has demonstrated that selective MEK1/2 inhibitor, trametinib, has a potential inhibitory effect on LPS-mediated inflammation in macrophages.50 These results suggest that MEK inhibitor can target not only histological abnormalities of IECs but also aberrant intestinal inflammation.
Here, we present a rational strategy for CD based on a systematic drug repositioning bioinformatics approach using a publicly available patient-derived gene signature to explore connections with existing drugs. One might assume that effective clinical treatments serve not only to reverse the clinical course of the disease but also to restore disease-affected molecular-level phenotypes to their normal level. Notably, several recent studies have made this assumption by directly searching for drug therapies whose molecular effects anticorrelate with disease molecular signatures using a CMap-based computational approach.26, 51–54 Although drugs were validated in preclinical models, it remains unknown whether the disease gene expression was reversed in disease models or not. It is important to note the limitations of this study. The gene signature, which was extracted from patients, can be provided as a “snapshot” in remission. Therefore, the interpretation of the “snapshot” requires careful thought about the result without excessive speculation. Moreover, some genomic effects might be agent specific; thus further studies will be needed to compare clinical outcomes with different anti-inflammatory therapeutic agents.
Nevertheless, here we showed that MEK1/2 inhibitor could significantly reverse the residual TUG signature to a normal state in gene expression in vitro. Also, colon-targeted delivery of MEK1/2 inhibitor was associated with both the successful improvement of diarrhea and restoration of histological abnormalities in vivo. These results suggest that additional clinical investigation using MEK1/2 inhibitor could be beneficial, especially contributing to accelerating restitution of dysregulated IECs and to preventing recurrence as a solution to unmet medical needs in CD therapy. In practice, however, there is no direct evidence showing how sustainable the clinical and endoscopic effectiveness of biological therapies for CD are. Hence, a close study focusing not only on effectiveness but also on large-scale long-term safety in a clinical setting is necessary for our purpose. In addition, a colon-targeting approach could ensure direct treatment at the inflamed site with lower dosing, and hopefully lead to more optimal and less toxic promising therapeutic strategies in the future. Finally, our study provides a sound rationale for predicting a drug’s potential therapeutic effect, which is based on its ability to induce molecular remission. It would exert the most favorable clinical outcomes rather than modulating only a particular subset of disease phenotypes.
SUPPLEMENTARY DATA
Supplementary data are available at Inflammatory Bowel Diseases online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoshiki Katou and Hikaru Saito for technical assistance; Kuniko Kikuchi and Hirokazu Tozaki for conducting next-generation sequencing (NGS); Tomoya Yata and Ayako Baba for providing microparticle formulation; Ikumi Chisaki for conducting pharmacokinetic studies; Shinya Tasaki for helpful advice regarding NGS data analysis; Yusuke Nakayama for establishment of an internal in silico screening system; and Christopher Arendt and Masayuki Ii for their guidance and support during the course of this work.
Conflicts of Interest: No potential conflicts of interest were disclosed.
REFERENCES
- 1. Lichtenstein GR, Abreu MT, Cohen R et al. ; American Gastroenterological Association American Gastroenterological Association Institute medical position statement on corticosteroids, immunomodulators, and infliximab in inflammatory bowel disease. Gastroenterology. 2006;130:935–9. [DOI] [PubMed] [Google Scholar]
- 2. Rutgeerts P, D’Haens G, Targan S et al. Efficacy and safety of retreatment with anti-tumor necrosis factor antibody (infliximab) to maintain remission in Crohn’s disease. Gastroenterology. 1999;117:761–9. [DOI] [PubMed] [Google Scholar]
- 3. Lakatos PL, Golovics PA, David G et al. Has there been a change in the natural history of Crohn’s disease? Surgical rates and medical management in a population-based inception cohort from Western Hungary between 1977-2009. Am J Gastroenterol. 2012;107:579–88. [DOI] [PubMed] [Google Scholar]
- 4. Lazarev M, Ullman T, Schraut WH et al. Small bowel resection rates in Crohn’s disease and the indication for surgery over time: experience from a large tertiary care center. Inflamm Bowel Dis. 2010;16:830–5. [DOI] [PubMed] [Google Scholar]
- 5. Peyrin-Biroulet L, Loftus EV Jr, Colombel JF et al. The natural history of adult Crohn’s disease in population-based cohorts. Am J Gastroenterol. 2010;105:289–97. [DOI] [PubMed] [Google Scholar]
- 6. Gisbert JP, Marín AC, Chaparro M. The risk of relapse after anti-TNF discontinuation in inflammatory bowel disease: systematic review and meta-analysis. Am J Gastroenterol. 2016;111:632–47. [DOI] [PubMed] [Google Scholar]
- 7. Boyapati R, Satsangi J, Ho GT. Pathogenesis of Crohn’s disease. F1000prime Rep. 2015;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Talley NJ, Abreu MT, Achkar JP et al. An evidence-based systematic review on medical therapies for inflammatory bowel disease. Am J Gastroenterol. 2011;106(Suppl 1):S2–S25; quiz S26 [DOI] [PubMed] [Google Scholar]
- 9. Zallot C, Peyrin-Biroulet L. Deep remission in inflammatory bowel disease: looking beyond symptoms. Curr Gastroenterol Rep. 2013;15:315. [DOI] [PubMed] [Google Scholar]
- 10. Kiesslich R, Duckworth CA, Moussata D et al. Local barrier dysfunction identified by confocal laser endomicroscopy predicts relapse in inflammatory bowel disease. Gut. 2012;61:1146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. D’haens G, Van Deventer S, Van Hogezand R et al. Endoscopic and histological healing with infliximab anti-tumor necrosis factor antibodies in Crohn’s disease: a European multicenter trial. Gastroenterology. 1999;116:1029–34. [DOI] [PubMed] [Google Scholar]
- 12. Bai JP, Alekseyenko AV, Statnikov A et al. Strategic applications of gene expression: from drug discovery/development to bedside. Aaps J. 2013;15:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen B, Butte AJ. Leveraging big data to transform target selection and drug discovery. Clin Pharmacol Ther. 2016;99:285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamb J, Crawford ED, Peck D et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. [DOI] [PubMed] [Google Scholar]
- 15. Cheng J, Yang L, Kumar V et al. Systematic evaluation of connectivity map for disease indications. Genome Med. 2014;6:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arijs I, De Hertogh G, Lemaire K et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;4:e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D’Haens GR, Geboes K, Peeters M et al. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–7. [DOI] [PubMed] [Google Scholar]
- 18. Rutgeerts P, Sandborn WJ, Feagan BG et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462–76. [DOI] [PubMed] [Google Scholar]
- 19. Geboes K, Riddell R, Ost A et al. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut. 2000;47:404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jung P, Sato T, Merlos-Suárez A et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–7. [DOI] [PubMed] [Google Scholar]
- 21. Merlos-Suárez A, Barriga FM, Jung P et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–24. [DOI] [PubMed] [Google Scholar]
- 22. Adams ME, Wallace MB, Kanouni T et al. Design and synthesis of orally available MEK inhibitors with potent in vivo antitumor efficacy. Bioorg Med Chem Lett. 2012;22:2411–4. [DOI] [PubMed] [Google Scholar]
- 23. Alhnan MA, Basit AW. Engineering polymer blend microparticles: an investigation into the influence of polymer blend distribution and interaction. Eur J Pharm Sci. 2011;42:30–6. [DOI] [PubMed] [Google Scholar]
- 24. Kendall RA, Alhnan MA, Nilkumhang S et al. Fabrication and in vivo evaluation of highly ph-responsive acrylic microparticles for targeted gastrointestinal delivery. Eur J Pharm Sci. 2009;37:284–90. [DOI] [PubMed] [Google Scholar]
- 25. Melé M, Ferreira PG, Reverter F et al. ; GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dudley JT, Sirota M, Shenoy M et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dry JR, Pavey S, Pratilas CA et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res. 2010;70:2264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sääf AM, Halbleib JM, Chen X et al. Parallels between global transcriptional programs of polarizing caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell. 2007;18:4245–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bregenholt S, Reimann J, Claesson MH. Proliferation and apoptosis of lamina propria CD4+ T cells from scid mice with inflammatory bowel disease. Eur J Immunol. 1998;28:3655–63. [DOI] [PubMed] [Google Scholar]
- 30. Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11:385–400. [DOI] [PubMed] [Google Scholar]
- 31. Hua S, Marks E, Schneider JJ et al. Advances in oral nano-delivery systems for colon targeted drug delivery in inflammatory bowel disease: selective targeting to diseased versus healthy tissue. Nanomedicine. 2015;11:1117–32. [DOI] [PubMed] [Google Scholar]
- 32. Lapidus A, Bernell O, Hellers G et al. Clinical course of colorectal Crohn’s disease: a 35-year follow-up study of 507 patients. Gastroenterology. 1998;114:1151–60. [DOI] [PubMed] [Google Scholar]
- 33. Chauvin A, Le Thuaut A, Belhassan M et al. Infliximab as a bridge to remission maintained by antimetabolite therapy in Crohn’s disease: a retrospective study. Dig Liver Dis. 2014;46:695–700. [DOI] [PubMed] [Google Scholar]
- 34. Coskun M. Intestinal epithelium in inflammatory bowel disease. Front Med (Lausanne). 2014;1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–53. [DOI] [PubMed] [Google Scholar]
- 36. Pastorelli L, De Salvo C, Mercado JR et al. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koch S, Nusrat A. The life and death of epithelia during inflammation: lessons learned from the gut. Annu Rev Pathol. 2012;7:35–60. [DOI] [PubMed] [Google Scholar]
- 38. D’Incà R, Di Leo V, Corrao G et al. Intestinal permeability test as a predictor of clinical course in Crohn’s disease. Am J Gastroenterol. 1999;94:2956–60. [DOI] [PubMed] [Google Scholar]
- 39. Arnott ID, Kingstone K, Ghosh S. Abnormal intestinal permeability predicts relapse in inactive Crohn disease. Scand J Gastroenterol. 2000;35:1163–9. [DOI] [PubMed] [Google Scholar]
- 40. Vereecke L, Beyaert R, van Loo G. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol Med. 2011;17:584–93. [DOI] [PubMed] [Google Scholar]
- 41. Suárez-Fariñas M, Fuentes-Duculan J, Lowes MA et al. Resolved psoriasis lesions retain expression of a subset of disease-related genes. J Invest Dermatol. 2011;131:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Planell N, Lozano JJ, Mora-Buch R et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62:967–76. [DOI] [PubMed] [Google Scholar]
- 43. Zeissig S, Bergann T, Fromm A et al. Altered enac expression leads to impaired sodium absorption in the noninflamed intestine in Crohn’s disease. Gastroenterology. 2008;134:1436–47. [DOI] [PubMed] [Google Scholar]
- 44. Herr R, Köhler M, Andrlová H et al. B-raf inhibitors induce epithelial differentiation in BRAF-mutant colorectal cancer cells. Cancer Res. 2015;75:216–29. [DOI] [PubMed] [Google Scholar]
- 45. Lemieux E, Boucher MJ, Mongrain S et al. Constitutive activation of the MEK/ERK pathway inhibits intestinal epithelial cell differentiation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G719–G730. [DOI] [PubMed] [Google Scholar]
- 46. Coskun M. The role of CDX2 in inflammatory bowel disease. Dan Med J. 2014;61:B4820. [PubMed] [Google Scholar]
- 47. Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dedhia PH, Bertaux-Skeirik N, Zavros Y et al. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150:1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu F, Huang J, Ning B et al. Drug discovery via human-derived stem cell organoids. Front Pharmacol. 2016;7:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Du SL, Yuan X, Zhan S et al. Trametinib, a novel MEK kinase inhibitor, suppresses lipopolysaccharide-induced tumor necrosis factor (TNF)-α production and endotoxin shock. Biochem Biophys Res Commun. 2015;458:667–73. [DOI] [PubMed] [Google Scholar]
- 51. Sirota M, Dudley JT, Kim J et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jahchan NS, Dudley JT, Mazur PK et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zerbini LF, Bhasin MK, de Vasconcellos JF et al. Computational repositioning and preclinical validation of pentamidine for renal cell cancer. Mol Cancer Ther. 2014;13:1929–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Claerhout S, Lim JY, Choi W et al. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6:e24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hirose M, Okaniwa M, Hayashi Y et al. Preparation of heterocyclic compounds as Raf inhibitors for treatment of cancer. WIPO patent WO2009028629 A1. March 5, 2009. [Google Scholar]
- 56. Ishikawa T, Banno H, Kawakita Y et al. Preparation of fused heterocyclic compounds as tyrosine kinase inhibitors. WIPO patent WO2008072634 A1. June 19, 2008. [Google Scholar]
- 57. Kawakita Y, Miwa K, Seto M et al. Design and synthesis of pyrrolo[3,2-d]pyrimidine HER2/EGFR dual inhibitors: improvement of the physicochemical and pharmacokinetic profiles for potent in vivo anti-tumor efficacy. Bioorg Med Chem. 2012;20:6171–80. [DOI] [PubMed] [Google Scholar]
- 58. Hirose M, Okaniwa M, Miyazaki T et al. Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors: 3. Evaluation of 5-amino-linked thiazolo[5,4-d]pyrimidine and thiazolo[5,4-b]pyridine derivatives. Bioorg Med Chem. 2012;20:5600–15. [DOI] [PubMed] [Google Scholar]
- 59. Barrett SD, Bridges AJ, Dudley DT et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18:6501–04. [DOI] [PubMed] [Google Scholar]
- 60. Mini E, Nobili S, Caciagli B et al. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17(Suppl 5):v7–12. [DOI] [PubMed] [Google Scholar]
- 61. Dong Q, Feher V, Kaldor SW et al. MEK1/2 inhibitors, methods for their synthesis, and their use in disease treatment. US patent US20080125437 A1. May 29, 2008. [Google Scholar]
- 62. Ishikawa T, Seto M, Banno H et al. Design and synthesis of novel human epidermal growth factor receptor 2 (HER2)/epidermal growth factor receptor (EGFR) dual inhibitors bearing a pyrrolo[3,2-d]pyrimidine scaffold. J Med Chem. 2011;54:8030–50. [DOI] [PubMed] [Google Scholar]
- 63. Dong Q, Dougan DR, Gong X et al. Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer. Bioorg Med Chem Lett. 2011;21:1315–9. [DOI] [PubMed] [Google Scholar]
- 64. Schellhammer PF. An evaluation of bicalutamide in the treatment of prostate cancer. Expert Opin Pharmacother. 2002;3:1313–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data of our study have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). The Ampli-seq data of LoVo cell line have been deposited under accession number GSE108050, and the RNA-seq data of murine colitis model have been deposited under accession number GSE108052.