Figure 1.
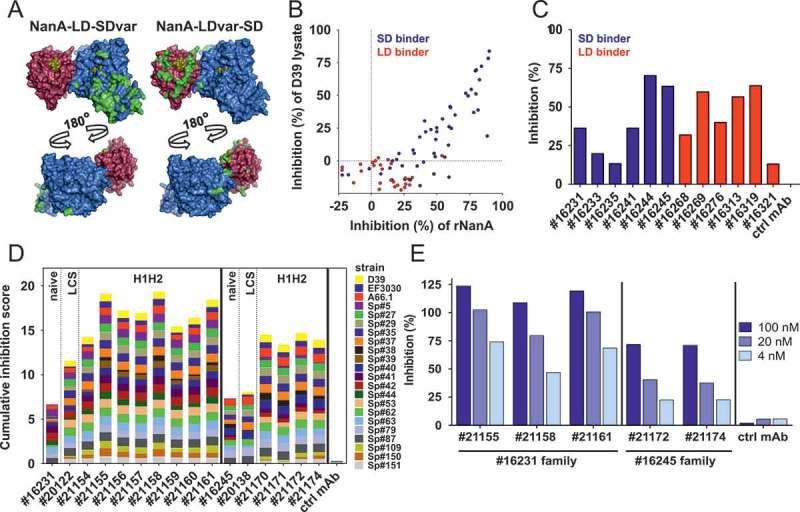
Generation of α-NanA mAbs. (A) Sialidase (blue, PDB 3H72 [26]) and lectin (red, PDB 4C1X [66]) domains of NanA aligned (using PyMOL software v2.0.5; Schrödinger) on the NanB structure (PDB 2VW1 [25]) as a scaffold. Residues differing from NanA-LD-SD are highlighted in green, and sugars occupying the sugar binding sites are shown in yellow. (B) Inhibition of neuraminidase activity of recombinant NanA (2.5 nM) and NanA from D39 lysates (used at a total protein concentration of 20 μg/mL) by naïve yeast library derived α-NanA mAbs (1 or 0.33 µM, respectively) in a fluorometric assay. (C) Inhibition of 1 nM rNanA-mediated Raji cell surface desialylation by naïve α-NanA mAbs (1 µM). (D) Cumulative inhibition of NanA sialidase activity in pneumococcal lysates (20–40 μg/mL) by selected naïve, and LCS (light chain shuffled) or mutated HCDR1 + HCDR2 (H1H2) SD-binding mAbs (0.33 µM) in a fluorometric assay. Inhibition scores for each lysate from 0 (no inhibition) to 1 (100% inhibition) were calculated relative to a control human IgG1. Cumulative inhibition scores were generated by addition of individual values (minimum score = 0, maximum score = 22). (E) Inhibition of Raji cell surface desialylation during infection with S. pneumoniae (D39Δply) by H1H2 α-NanA mAbs at the indicated concentrations. One representative experiment from the original screening data is shown in panels B, C and E. Two independent experiments are summarized in panel D.
