Abstract
Type 1 diabetes (T1D) culminates in the autoimmune destruction of the pancreatic β-cells, leading to insufficient production of insulin and development of hyperglycemia. Serum biomarkers including a combination of glucose, glycated molecules, c-peptide, and autoantibodies have been well established for the diagnosis of T1D. However, these molecules often mark a late stage of the disease when ~90% of the pancreatic insulin-producing β-cells have already been lost. With the prevalence of T1D increasing worldwide and because of the physical and psychological burden induced by this disease, there is a great need for prognostic biomarkers to predict T1D development or progression. This would allow us to identify individuals at high risk for early prevention and intervention. Therefore, considerable efforts have been dedicated to the understanding of disease etiology and the discovery of novel biomarkers in the last few decades. The advent of high-throughput and sensitive ‘-omics’ technologies for the study of proteins, nucleic acids, and metabolites have allowed large scale profiling of protein expression and gene changes in T1D patients relative to disease-free controls. In this review, we briefly discuss the classical diagnostic biomarkers of T1D but mainly focus on the novel biomarkers that are identified as markers of β-cell destruction and screened with the use of state-of-the-art ‘-omics’ technologies.
1. Introduction
Type 1 diabetes (T1D) is a chronic autoimmune disease characterized by insulin deficiency as a consequence of autoimmune-mediated destruction of insulin producing pancreatic β-celIs within the islets of Langerhans. Although a lifelong administration of exogenous insulin can help to balance glucose homeostasis in T1D patients to a certain degree, currently there are no effective curative therapies available for this disease. At the time of diagnosis, 80% to 90% of β-cell mass has been lost[1] and it is generally accepted that it would be extremely difficult to intervene or reverse the progression of T1D at this late stage.[2] Alarmingly, a steady increase in the number of diagnoses of T1D has been observed, with an average annual increase of 2–5% worldwide.[3] The age of symptomatic onset is usually during childhood or adolescent with a peak incidence rate at 12–14 years of age, but the symptoms can also develop at much later ages [4]. Moreover, T1D is often accompanied by sudden and acute complications. Concerns about their future health could lead to development of practical and emotional problems for patients and their families.[5,6] All of these concerns urgently demand more efficient means for early prediction, monitoring of progression, and eventually prevention or reversal of the disease especially at an early stage.
Biomarkers are indicators of normal and abnormal physiological or pathological processes and serve important roles in clinical diagnosis, prognosis, as well as monitoring therapeutic responses. The development of specific blood serum (or plasma) biomarkers are particularly attractive for most diseases because of the nature of blood circulation throughout the whole body, which serves as a carrier of molecules which were changed at localized pathological sites and are relatively easy to access compared to other biological fluids or pathological tissues.[7] However, the development of specific serum biomarkers, especially those reflecting pancreatic β-cell death or stress, has been challenging for T1D because of the fact that T1D is the result of autoimmune attack of β-cel Is, which are only about 0.002% of body mass.[8] Although T1D has been investigated for over a century, the etiology of the disease is still not fully understood. Until now, the biomarkers implemented in clinical practice such as glucose, HbAlc, c-peptide, and autoantibodies (AAb) were mainly diagnostic markers, although AAbs serve as relatively good prognostic markers of the risk of eventual development of the disease. While there has been advances in our understanding of the pathogenesis of T1D[9], we still do not have effective serum markers that can reflect the β-cell function, stress, β-cell mass, or serve as predictors of the progression of disease development. In this review, we will briefly discuss how these traditional biomarkers are used in the clinical diagnosis of T1D followed by the utility of other biomolecules which have resulted in a better understanding of T1D etiology. The contribution of advanced ‘-omics’ technologies leading to the discovery of potential serum biomarkers will also be discussed. Finally, we will discuss the potential biomarkers which can be used to predict disease, including the use of the best currently established AAb-based biomarkers and other novel predictive candidate biomarkers. Figure 1 shows the major types of potential serum biomarkers for the diagnosis and prediction of T1D.
Figure 1. The many types of currently used and potential biomarkers from serum for the diagnosis and prediction of T1D.
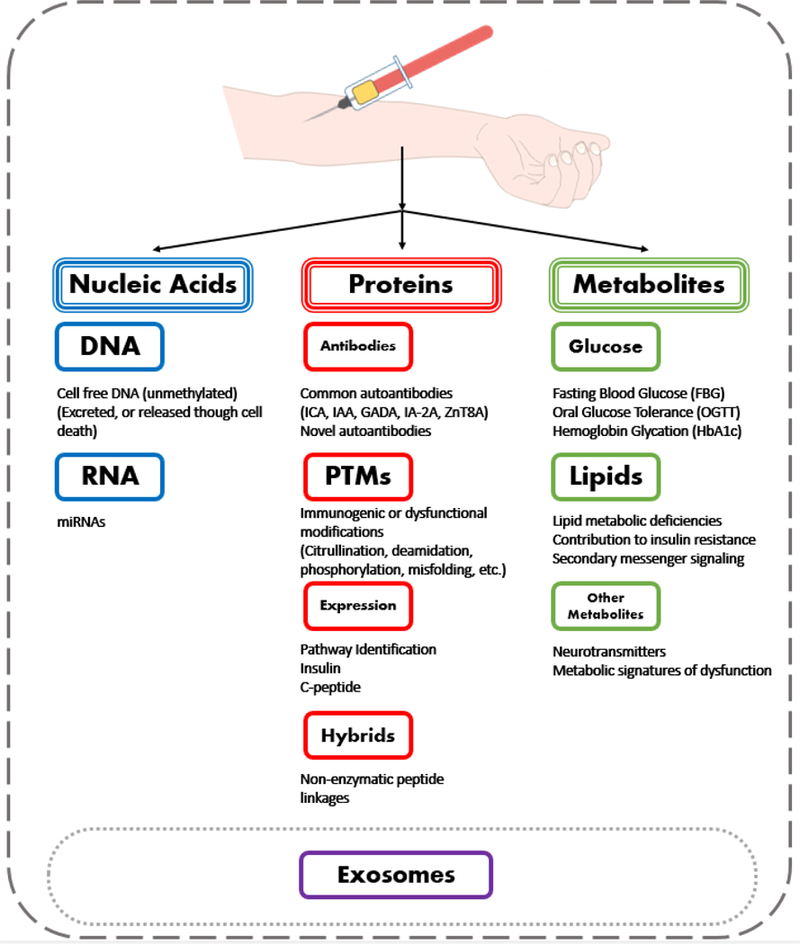
Types of validated clinical diagnostic biomarkers and potential new types of biomarkers are illustrated.
2. Serum diagnostic markers
Development of T1D involves many factors including genetics, environmental triggering and modifying factors. However, none of these factors can serve as a clear criterion for diagnosis. The current diagnostic biomarkers of T1D still rely on the consequences of hyperglycemia, such as the resulting high glucose or glycated hemoglobin, in combination with other T1D specific biomarkers that discern T1D from other subtypes of diabetes, such as low C-peptide levels or AAbs.
2.1. Glucose-related biomarkers
Diabetes may be diagnosed with a random plasma glucose ≥ 200 mg/dL in patients with classic symptoms of hyperglycemia or hyperglycemic crisis, or two repeated positive results of glucose-related tests in the absence of unequivocal hyperglycemia. In the latter case, the blood-based glucose-related testing includes hemoglobin Ale (HbAlc) (a product of non-enzymatic glycation), the fasting plasma glucose (FPG), and oral glucose tolerance test (OGTT). Each of these tests has its own criteria, sensitivity, and specificity for the diagnosis of diabetes. The American Diabetes Association (ADA) criteria for diagnosis of diabetes is HbAlc ≥ 6.5%, or FPG ≥ 126 mg/dL, or 2-hour plasma glucose of OGTT ≥ 200 mg/dL. HbAlc test provides an indirect measure of the average blood glucose for the past 2 to 3 months, which is a reflection of chronic hyperglycemia, but it is not as sensitive as FPG and OGTT in diabetes diagnosis. FPG and especially OGTT results can be more variable and a close follow-up repeated test is often necessary.
2.2. Autoantibody-based biomarkers
Of the many types and subtypes of diabetes only 5–10% are classified as T1D. They usually can be differentiated from type 2 diabetes (T2D) and other subtypes based on the presence of specific AAbs. There are five commonly tested AAb markers used in the diagnosis of T1D[10–13] which include ICA (islet-cell cytoplasmic AAb), GADA (glutamic acid decarboxylase (GAD) AAb), IA-2A (insulinoma 2 (IA-2)- associated AAb), IAA (insulin AAb), ZNT8A (zinc transporter 8 AAb). At least 1 autoantibody is present in > 95% of individuals with T1D upon hyperglycemia detection.[14,15] Despite being identified through validated highly confident assays, the commonly tested AAbs have been shown to be insufficient for detection of all T1D cases as there remains a subset of patients who do not have the above listed AAbs at diagnosis, indicating either a possibility of insensitive tests or the potential of different types of autoantigen-mediated β-cell destruction in these T1D patients.
2.3. C-peptide
Insulin is first synthesized as preproinsulin which becomes proinsulin after cleavage of the signal peptide in the endoplasmic reticulum. Proinsulin is packaged into vesicles and is then cleaved into the active insulin hormone and C-peptide and both insulin and C-peptide are released in equimolar amounts from mature granules, together with small amounts of uncleaved proinsulin.[16] Due to hepatic uptake, insulin has a much shorter half-life time (3–5 min) compared to proinsulin and C-peptide. Stimulated serum C-peptide level (as a surrogate for insulin) has been considered a consistent and sensitive measure of β-cell function and can be used to help differentiate autoimmune-diabetes from other diabetes subtypes[17] With the discovery of more and highly prevalent complications, C-peptide test needs to be combined with other factors, such as BMI, for better interpretation of disease progression.[18] C-peptide measurement is also currently the most suitable primary outcome for clinical trials of therapies aimed at preserving or improving endogenous insulin secretion in T1D patients.[19] However, C-peptide measurement may not be as reliable with the increasing obesity epidemic.
When diagnosing diabetes in the general population, it is most common to detect and evaluate hyperglycemia and blood glucose-related markers. In combination with AAb testing, T1D can be typically differentiated from other forms of diabetes. However, the manifestation of clinical T1D represents the near end-stage of β-cell destruction since only 10–20% of the insulin producing β-cells have been estimated to still be functioning at the time of diagnosis.[20] Therefore, it is imperative to have specific markers that can monitor and predict the disease progression.
3. Predictive biomarkers for T1D development
Prior to clinical T1D, the disease is typically preceded by an asymptomatic period of β-cell destruction that is highly variable in duration, ranging from months to decades.[15,21] It is important to study this silent period of autoimmune destruction of β-cells before the onset of clinical disc to gain understanding of its etiology and discover predictive or prognostic biomarkers 1 development or progression.
3.1. Autoantibodies
Autoantibodies (AAbs) are not only markers for diagnosis and disease classification, but also regarded as the current gold standard for the prediction of T1D development. It is still unclear what the exact role of islet AAbs is in T1D pathogenesis. Many consider it unlikely that islet AAbs are the cause of T1D but rather a reflection of disease progression or a secondary response. They provide proof of immune activation against certain autoantigens in the insulin producing β-cells in AAb individuals.
While the list of known autoantigen recognizing AAbs is still expanding, among all discovered AAbs, ICA, GADA, IAA, IA-2A and ZnT8A remain the most sensitive and specific, and are not difficult to measure thereby warranting their utility in T1D diagnosis and prognosis. The first-degree relatives (FDRs) of patients diagnosed with T1D or those having high-risk human leukocyte antigen (HLA) genotypes, are highly recommended to be screened for these AAbs to estimate the risk of T1D development. Seroconversion has a major impact on the accuracy of predicting T1D. After seroconversion to any two of the five common AAbs, an individual will almost always develop clinical T1D given sufficient time.[15] However the rate of progression depends on many factors, including type and number of AAbs, age, genotype, sex, fitness, etc.
Due to the heterogeneity of the disease, there is not a simple yes/no interpretation for T1D prediction. Autoantibody phenotype and numbers, epitopes, affinity, appearance order, combined with patients’ genotypes and ages all contribute to the T1D risks.[22] For example, IAA and GADA are more frequently detected as the first AAb in children but often disappear in 25% and 10% of cases respectively in children at clinical onset. While only a small fraction of the children had IA-2A or ZnT8A as their first AAb and they all persist through diagnosis.[15,23–26] Each of these antibodies also vary in their affinities and epitope specificities, and these variations are connected with different risks for T1D.[27] Therefore, it makes sense to include the AAb phenotype and epitopes when defining the T1D risks. However, individuals positive for a single islet AAb are far less likely to develop T1D than individuals who are positive for multiple islet AAbs. Progression to T1D in children with a single AAb is about 10% at 10 years, and the incidence of developing clinical T1D after more than 2 AAbs are present is 11% each year and >70% in the ensuing 10 years.[15,22,28] In general, the number of islet AAbs expressed by an individual is a more important predictor of T1D than any specific combination of islet AAbs.[29]
Despite the clear utility of the AAbs combined with genetic susceptibility in T1D prediction, they have several limitations.[30–32] First, AAb screening is only recommended for FDRs of T1D patients in the settings of clinical research studies[33] and more than 85% of people who develop T1D do not have family history of disease. Second, only a subset of the AAb-positive subjects will progress to clinical diabetes, all in different time frames, and the presence and levels of AAbs after seroconversion do not track disease progression. Therefore, it is critical to have additional biomarkers that can predict the stages of progression and identify the best timing for intervention and therapy. Finally, AAbs are also not useful as biomarkers for therapeutic outcomes. Consequently, in the last few decades, considerable efforts have been dedicated utilizing new technologies for the discovery of novel serum biomarkers that can serve as predictors of disease progression, β-cell function and mass, and functional monitoring of therapeutic responses.
3.2. Potential novel biomarkers
The development of high-throughput ‘-omics’ technologies has provided some excellent platforms for the discovery of novel biomarker candidates by allowing a systems-wide coverage of molecular changes during disease progression. Often these studies are conducted using cultured primary cells,[34–36] immortalized cells,[37] mouse islets,[38] or with human pancreas tissues[39] including small quantity islets isolated by laser microdissection,[40,41] single islet sections,[42] or sorted cell sources.[43,44]. While many of these studies may not directly involve serum, they are useful in identifying potential serum biomarker candidates while also elucidating dysregulated pathways underlying the disease. These methods can aid in identifying novel biomarker candidates potentially useful for indicating the level of β-cell destruction, dysfunction, and mass as well as the ongoing immunological response in serum. In this part, we mainly discuss three types of novel biomarkers including proteins, nucleic acids, and metabolites. Figure 2 illustrates the types of markers and the main omics technologies or assay platforms applied in the discovery and validation of these types of serum biomarkers in T1D.
Figure 2.
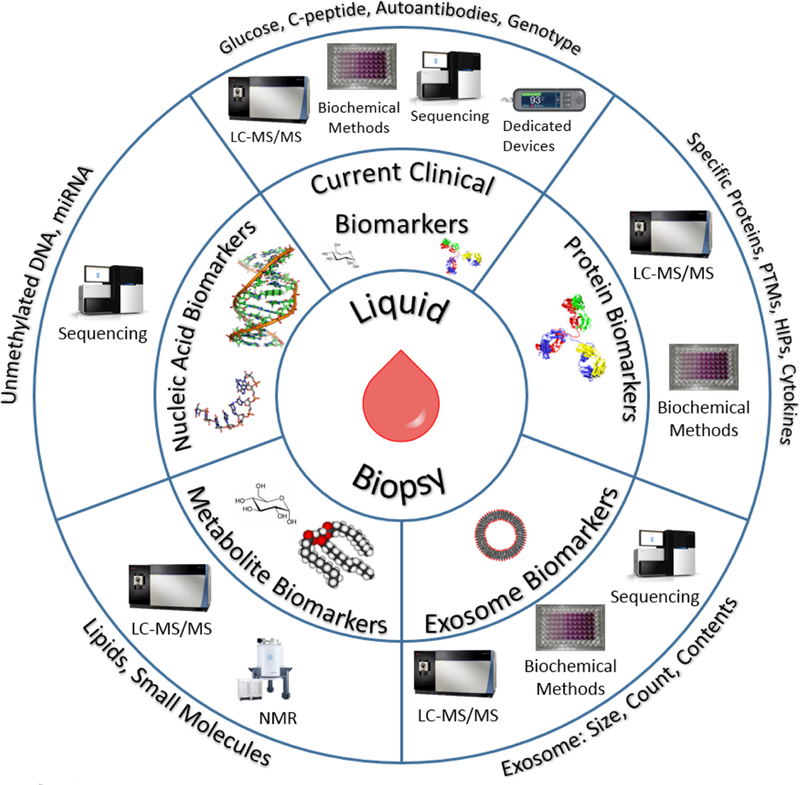
Different types of potential serum biomarkers for T1D and the associated omics technologies and assay platforms for their discovery and measurements.
3.2.1. Protein biomarkers
3.2.1.1. Serum protein biomarkers
Proteins are the direct executors for all aberrant genetic changes and the identification of specific serum proteins as the source of biomarkers for specific diseases perhaps is the most accepted concept in the biomarker field. Proteomics analyses of protein/peptide expression and post-translational modifications (PTMs) for T1D has the potential to make significant contributions to aid in disease prediction and prognosis. Comprehensive analysis of the serum proteins is generally challenging because of the extreme complexity and high dynamic range of the serum proteome. This difficulty is especially true considering the extreme low abundance of potential biomarkers from disease-specific tissues or cells such as pancreatic β-cells. Proteomics analysis of serum is also complicated by the heterogeneity of the disease, individual variations, different disease stages, and the often subtle yet significant abundance changes. Therefore, well-designed strategies that incorporate sensitive proteomics technologies and experimental designs with statistical considerations are critical for addressing each of the aforementioned barriers. Perhaps due to the extremely low mass of pancreatic β cells in total body mass and the long progressive nature of β-cell loss, the identification of serum protein biomarkers specific to T1D is still in its early stage. Herein, we will present a brief discussion of the current proteomics workflow for biomarker development and a summary of candidate protein biomarkers reported for T1D from recent literature.
A general proteomics workflow used for the discovery of novel serum biomarkers usually consists of two phases: a discovery phase using bottom-up (peptide centric) proteomics to search for biomarker candidates, and a verification phase of selected candidates using targeted proteomics analysis and/or immunoassays. The extremely high dynamic range of serum proteome often requires the removal or depletion of major high-abundance proteins using immobilized antibodies in order to enhance the detection of low-abundance candidate biomarkers.[45,46] While such strategies make the low-abundance proteins relatively accessible, they could inadvertently remove proteins of possible interest through binding (specific or nonspecific) to targeted high-abundance proteins. A second strategy to deal with the high dynamic range issue is extensive serum protein/peptide fractionation, usually with chromatography methods that are orthogonal to the final dimension of low-pH reverse phase liquid chromatography (RPLC) before the MS analysis.[47] Both of these strategies have been employed or combined to profile serum proteome to a depth of several hundreds to thousands of proteins[48–53] from people without and with prediabetes/diabetes. However, the extensive fractionation decreases the analytical throughput of sample analysis and thus limits the number of samples to be analyzed. The throughput can be partially recovered by multiplexing with unique sample specific mass tags such as iTRAQ[54] (isobaric tag for relative and absolute quantification) or TMT[55] (tandem mass tags) and this approach has been implemented in longitudinal proteomics profiling of human plasma.[52,53] Typically, potential protein biomarkers that differ between cases and controls with statistical significance are then followed by verification with targeted proteomics and/or immunoassay in an independent cohort of samples. Table 1 lists the biomarker candidates that were reported from at least two independent research papers. Unfortunately, most of these proteins are highly abundant in plasma and lack specificity to pancreatic β cells and the overlap between the different studies is limited. Moreover, some of them showed opposite changes in terms of up/down expression regulation in different studies. The observed inconsistency and current lack of highly promising protein biomarkers could be a result of the limitations of the shotgun proteomics approach in identifying low abundance proteins specific to pancreatic islet cells and the heterogeneity of disease and individuals masking the “true biomarkers”. More in-depth profiling with advanced technologies along with improved experiment designs (e.g., longitudinal samples) will be necessary to discover more specific protein biomarkers that show statistically significant changes during disease progression.[56]
Table 1.
Protein candidate biomarkers discovered by proteomics.
| Gene Symbol | Protein name | Regulation | Function |
|---|---|---|---|
| ADIPOQ | Adiponectin | Zhi et al,[48] up* Moudler et al,[52] down* |
Glucose homeostasis |
| APOA4 | Apolipoprotein A-IV | Zhi et al,[48] up von Tonerne, et al,[49] down Moudler et al,[52] down | Leukocyte adhesion |
| APOC4 | Apolipoprotein C-IV | von Tonerne, et al,[49] down Moudler et al,[52] down | Viral infection |
| AZGP1 | a-2-glycoprotein 1 (zinc) | Metz et al,[50] up Zhang et al,[51] up |
Lipid mobilization activity |
| BTD | Biotinidase | von Tonerne, et al,[49] up Zhang et al,[51] up | Biotinidase activity |
| C3 | Complement C3b | Zhi et al,[48] down von Tonerne, et al,[49] down Zhang et al,[51] down |
Innate immunity, complement activation |
| C4A | Complement C4-A | Zhi et al,[48] down von Tonerne, et al,[49] down Zhang et al,[51] down | Innate immunity, complement activation |
| CLU | Clusterin | von Tonerne, et al,[49] down Metz et al,[50] down Zhang et al,[51] down |
Cytoprotective capability |
| KNG1 | Kininogen 1 isoform 1 | Zhi et al,[48] up von Tonerne, et al,[49] down Zhang et al,[51] up | Innate immunity, DC activation |
| LUM | Lumican | Metz et al,[50] up Zhang et al,[51] up |
Extracellular matrix structural constituent |
| SERPINA6 | Corticosteroid-binding protein |
Metz et al,[50] up Zhang et al,[51] up |
Correlate to insulin deficiency or response |
| SERPINF2 | Alpha-2-antiplasmin | von Tonerne, et al,[49] down Zhang et al,[51] up | Serine protease inhibitor |
| TTR | transthyretin | von Tonerne, et al,[49] down Zhang et al,[51] down | Involve in β-cell stimulus- secretion coupling |
Up/down inc licates that the protein is significantly up/down regulated in serum of patients with iabetes diabetes/prec relative to the healthy controls.
3.2.1.2. PTMs
The mechanisms underlying the breach of immune tolerance to β-cell antigens is still poorly understood. A prominent role for inflammation in cross-talk between the β-cell and immune system has been demonstrated in the pathogenesis of T1D. Inflammation triggers β-cell oxidative and endoplasmic reticulum (ER) stress, and may lead to alternative splicing and misfolding of β-cell proteins as well as post-translational modifications (PTMs).[57,58] Being different from the native proteins, the modified format of autoantigens or neoantigens (i.e., antigens expressed under specific conditions, rather than ubiquitously) maybe recognized as foreign and result in a breakdown of tolerance. More importantly, the newly discovered antigens can potentially be used as therapeutic agents to suppress autoimmunity in those at risk for the development of T1D, as well as in those with established disease who received islet replacement or regeneration therapy. Additionally, antibodies against these neoantigens could be supplementary to the current T1D AAb panel for diagnosis and prediction, since 2–5% of patients who are diagnosed in the clinic with T1D are found to be negative for common AAbs.[59] Despite the great potential of neoantigens in helping predict and prevent T1D, investigation of neoantigens is still in its infancy.
Increased recognition of post-translationally modified autoantigens by autoreactive T and B cells has been observed in other human autoimmune diseases, including rheumatoid arthritis and celiac disease. The similar PTMs observed in T1D include citrullination and deamidation of GAD65,[60] citrullination of ER chaperone 78 kDa glucose-regulated protein (GRP78),[61] deamidation of proinsulin peptide[62], ROS modified collagen type II (CM),[63] oxidative insulin,[64] phosphorylation of peripherin,[65] and formation of vicinal disulfide bond between adjacent cysteine residues of insulin A-chain.[66] PTMs of autoantigens enhance the immune response and recognition by T cells by either increasing the binding affinity to major histocompatibility complex class II (MHC II) molecules[63] or by modifying amino acids at T-cell contact positions.[60,66] These modified antigens activate antigen recognition by T cells isolated from T1D donors. More impressively, T-cell clones have been shown to respond specifically to these modified antigens which have been detected in the blood of mouse and human donors with prediabetes. Other types of modifications not previously mentioned that could play a role in immunogenicity, as observed in T1D or other autoimmune diseases, include phosphorylation[67], lipidation[68], glycation[69], etc. Another type of recently discovered novel antigen is hybrid insulin peptides (HIPs) which result from covalent crosslinking of proinsulin fragments with other peptides present in β-cell secretory granules.[70] HIPs are detectable in mouse pancreas and CD4 T cells isolated from T1D donors mount a memory recognition response to these HIPs. The potential utility of specific PTM signatures of autoantigens in serum for predicting T1D is still in exploratory stage.
3.2.1.3. Cytokines
Another type of important protein biomarkers are cytokines and chemokines since they play significant roles in the stimulation, regulation, and intercellular signaling of immune cells, mediating insulitis and β-cell destruction.[71] Many cytokines are upregulated in prediabetes[72] and their roles as surrogate serum markers of disease have been investigated. A screening of 65 cytokines in a nested case-control study of 67 children revealed that 15 cytokines, chemokines, and growth factors were elevated in AAb positive subjects and two specific cytokines, IL-10 and IL-21, can even differentiate enterovirus infected groups.[73] Serum CXCL1 was demonstrated to discriminate T1D from T2D.[74] An impressive study screened 13 cytokines from two independent sample sets across thousands of subjects. Interestingly, they discovered four cytokines (IL8, IL-IRa, MCP-1, and MIP-Ιβ) were all downregulated in the serum of T1D patients. They suspected those cytokines may play protective roles against T1D but additional studies would be required to understand their mechanisms.[75] All these studies used well established immunoassays but did not show much overlap to other studied cytokines or their trend between different studies. Cytokines are powerful mediators of inflammation and microbial elimination. However, a potential problem for the use of cytokines as disease serum biomarkers is the lack of organ specificity to T1D since serum cytokines can change in response to injury and damage in all types of tissues and in many inflammation or immune-related diseases.[76,77]
3.2.1.4. Proinsulin/C-peptide ratio
An elevation in the ratio of proinsulin to C-peptide (measurement of insulin) is indicative of β-cell dysfunction and is primarily thought of as a reflection of alterations in insulin protein folding and processing that originate in the endoplasmic reticulum.[78,79] Data has shown that the elevated ratio of serum proinsulin to C-peptide, as an indication of β-cell ER dysfunction, precedes T1D onset especially in younger children. [80] Future studies measuring proinsulin to C-peptide ratios, combined with other biomarkers, are needed to demonstrate their utility in predicting the onset of T1D in the pre- symptomatic phase.[80,81]
3.2.1.5. Other novel AAbs and autoantigens
There have been a number of reports of novel or rarer types of autoantigens and associated AAbs in T1D, including tetraspanin-7 (a membrane glycoprotein),[82], islet cell surface antigen(ICSA),[83] islet of Langerhans regenerating protein-la (REGIA),[84] pancreatic duodenal homeobox 1 (PDX1),[85] chemokine (C-C motif) ligand 3 (CCL3)[86] protein disulfide isomerase (PDl),[87] golgin-160,[87] voltage-gated potassium channel (Kvl.3),[87] Rab GDP dissociation inhibitor beta (GDIβ),[59] amylase alpha-2A,[88] L-type voltage-gated calcium channel s(VGCCs),[89,90] aminoacyl-tRNA synthetase,[91] protein family members of commonly tested autoantigens GAD67[92] and ΙΑ−2β,[93] splicing variants of the commonly tested autoantigens[94–96], “ as well as to interesting protein/peptide fusion products. [70,97] More targel are likely to exist as has been demonstrated using risk assessment analyses based on total proteome measurements[98] and bioinformatics expression analyses[99].
3.2.2. Nucleic acid biomarkers
The presence of low-concentration small circulating DNA fragments in serum documented.[100] The DNA fragments are thought to be the result of apoptosis, necrosis, or through active and as-yet-unknown secretion pathways.[101] These nucleic acids might carry a variety of genetic and epigenetic alternations related to disease development and progression. Recently, circulating unmethylated insulin DNA has aroused great interest as a biomarker target for early detection of β-cell death in T1D.[102,103] This is based on the fact that certain cytosine-guanine (CpG) sites in the insulin gene are specifically unmethylated in pancreatic β-cells and methylated in most other tissues. During progression of T1D, fragments of the characteristic unmethylated insulin DNA due to β-cell death are leaked into the bloodstream and become detectable. Therefore, measurement of the amount of unmethylated insulin DNA in the peripheral circulation may reflect the amount of β-cell death.[104] In addition, the new methylation-specific real-time PCR and sequencing techniques have proven to be sensitive, specific, and cost-effective assays[104–107] for identifying and quantifying unmethylated insulin DNA in blood.
Herold et al. led the screening of serum unmethylated insulin DNA as T1D biomarkers in several different cohorts and demonstrated the ratio of unmethylated/methylated INS DNA was consistently and significantly higher in patients with recent onset T1D compared to healthy groups, patients with long standing T1D, and the at-risk population with at least one AAb and normal OGTT.[103,104,106,108] The receiver operating characteristic (ROC) analysis revealed the ratio of unmethylated/methylated insulin DNA allows for discrimination between T1D and healthy groups with a sensitivity of 38% and a specificity of 95%.[106] Another recent work by Fisher et al. they reported the absolute levels of both unmethylated and methylated insulin DNA were higher in new onset T1D patients compared to control individuals and emphasized methylated insulin DNA may also be informative of the underlying disease process.[107] Investigation of unmethylated circulating DNA as a T1D biomarker is not restricted to insulin. Amylin is a glucose regulating hormone highly expressed and co-secreted with insulin from β-cells and interestingly, its unmethylated DNA is correlated with unmethylated insulin DNA as well in recent onset T1D and healthy subjects.[109] However, the utility of circulating DNA markers in T1D diagnosis and prognosis still needs to be further validated.
Another type of important nucleic acid biomarkers is called the transcriptomic fingerprint. Transcriptomic profiles in T1D have also been studied in samples of whole-blood,[110,111] peripheral blood mononuclear cells (PBMCs),[112] and other blood derived immunocyte subsets.[113] The suitability of the sample types used for transcriptional signatures has yet to be rigorously evaluated and the cellular composition of analyzed samples should be considered in the interpretation of the blood-based transcriptional data. A genome-wide transcriptomics analysis of whole blood RNA samples revealed the type 1 interferon (IFN) related transcriptomics signatures were already detectable in children at increased risk of T1D before the T1D-associated autoantibodies were detected.[110] This upregulation of IFN-inducible genes were found to be transient in isolated PBMCs from a different longitudinal cohort of children with a genetic predisposition to T1D.[112] Mehdi et al., combined the gene expression data from these studies and corrected it for time of seroconversion. This sophisticated statistical analysis identified differentially expressed genes that contribute to T cell-, DC-, and B cell-related immune response.
MicroRNAs (miRNAs) are a class of small (21–23 nucleotides) non-coding RNAs that are generally considered as regulators of gene expression through targeting of mRNAs or aiding mRNA stability. In recent years, new roles for miRNAs have been shown both in the regulation of β-cell function and the pathogenesis of T1D. A recent review reported that more than 200 dysregulated miRNAs have been identified in different tissues, cells, and serum/plasma in human and murine samples of T1D.[114] Among the miRNAs that were dysregulated in serum from human samples, several of them have the potentials of being used as circulating biomarkers of T1D. Increased serum levels of miR-375 have been considered to be linked with β-cell injury and significant increase of circulating miR-375 levels has been measured in subjects with T1D[115] and T2D.[116] However, Latrielle et al. showed that β-cell derived miR-375 only contributes to about 1% of the total serum miR-375 and questioned its capability of providing insights into β-cell loss.[117] Comparable or even decreased levels of miR-375 were also reported in disease cases.[118,119] Osipova et al. investigated circulating levels of three specific miRNAs using qPCR techniques and revealed that miR-21 and miR-210 were significantly increased in serum of subjects with T1D.[120] Similar to the discovery of proteomics biomarkers, global microarray and sequencing analysis of the serum miRNA for discovery and qPCR for validation were employed to screen miRNA biomarkers in T1D and healthy control subjects.[121] Among these analyzed miRNAs, miR-21,[115,120,121] miR-24,[115,121,122] miR-148a,[115,121] miR-181a-5p,[121,123] miR-210–5p,[120,121] are shown to be more consistently upregulated in serum from T1D cases in at least two studies.
MiRNAs have also been analyzed in blood derived exosome samples since exosomes are found to be the transporter of miRNAs in cell-to-cell communication in vitro.[124] Lakhter et al. found that serum exosomal miR-21–5p was increased in new onset T1D patients compared with healthy individuals while total serum miR-21–5p were reduced, suggesting that serum exosome miR-21–5p maybe a promising marker of T1D development.[125] Garcia-Contreras et al. discovered 7 differently regulated exosomal miRNAs in 12 pairs of case-control subjects using microarray techniques and only 3 of them had significant changes in a different cohort by qRT-PCR.[126] The authors suggested that this could be related to the differences of sensitivity and efficiency of the techniques and different disease status in the study population.
3.2.3. Metabolomics biomarkers
Glucose is a simple and useful metabolic biomarker used for the diagnosis of diabetes, but it is not specific to T1D. Research focusing on serum metabolomics biomarkers (comprising metabolites, lipids, and small molecules) is still in its infancy but is becoming more studied. Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry are the two techniques used to profile serum metabolic fingerprint of T1D.[127,128]. A few amino acids and lipid metabolites were found to be associated with T1D. Methionine is one typical amino acid that is involved in DNA methylation and could be relevant to the timing of appearance of autoantibodies.[127] Lipidomics studies of young and at-risk patients that progressed to clinical disease revealed that some classes of lipids show dysregulation in the blood.[129] Sphingolipids play important roles in inflammation and insulin signaling and have been analyzed for biomarker utility.[130] Torre et al. demonstrated that phospholipids were decreased in cord blood of T1D patients at the time of birth, long before diagnosis.[131] The lipids most decreased were phosphatidylcholines and phosphatidylethanolamines. A particularly interesting aspect of the study was the strong correlation of known cases of first trimester gestational infection and the corresponding decrease in measured phospholipids at birth, an observation made from several studies.[131–133] It has been proposed that low levels of phospholipids at birth may represent a higher risk factor for developing T1D due to the important role phospholipids play in immune system development and modulation. With further advances in discovery and validation efforts, metabolomics biomarkers has pot environmental factors contributing to the disease and in environmental factors contributing to the disease and in predicting the progression of the disease.
4. Future directions
While the diagnosis, or even prognosis, of T1D based on genotyping and the presence of multiple AAbs has been well established, it is still extremely challenging to develop specific biomarkers for predicting T1D development or progression. Considerable progress has been made in the application of various ‘-omics’ technologies toward the identification and validation of different kinds of biomarkers. Some of the biomarkers such as the methylation patterns of circulating DNA[103,105] have shown promising aspects in the detection of β-cell death and predicting of T1D development. Nevertheless, there are still many hurdles on the way to find novel biomarkers that will be specific to pancreatic β-cells in terms of β-cell function, stress, or death. It is important that the biomarkers are capable of detecting T1D development at very early stage, predicting the disease progression, stratifying high-risk population for intervention, and monitoring the efficacy of novel therapies.
While the most significant advances to date have occurred in the area of nucleic acid-based predictive biomarkers,[103,105,107] the direction of developing of novel β-cell specific proteins or PTM biomarkers along with immune-responsive markers for predicting T1D are still very attractive because proteins/PTMs (e.g. C-peptide) are more likely to provide an assessment of β-cell function or dysfunction. However, proteomics based discovery of β-cell specific markers has been challenged by the complexity of the serum/plasma sample, the extreme low-abundance of the candidates, and the sensitivity of the existing technologies. However, we anticipate that the currently used advanced proteomics technologies[134] should be sufficiently sensitive to detect and validate β-cell specific biomarkers in serum with well-designed sample cohorts. Moreover, the quality of the sample sources utilized for biomarker development is another critical point for success. The sample sets need to provide sufficient statistical power for candidate discovery and validation in order to establish the specificity and sensitivity. In order to develop a biomarker capable of accurately predicting progression, longitudinal samples from clinical trials such as T1D TrialNet for both high-risk subjects that progressed to T1D and those that did not progress to T1D will be important for such development.
Finally, besides serum/plasma, circulatory exosomes or extracellular vesicles may also serve as an interesting source for biomarkers in T1D.[135–137] Several studies have shown that β-cells, under conditions of insulitis, release exosomes to antigen presenting cells (APCs) which can lead to their activation. These exosomes contain several well documented auto-antigens including GAD65, IA-2, and proinsulin.[34] Conditions within exosomes may also cause differential modifications of the contents[138] and impaired immune phenotyping.[139] It is also possible that there isn’t a single ideal predictive biomarker. Instead multiple biomarkers may constitute a multi-marker panel of proteins, PTMs, or nucleic acids from serum and/or serum exosomes that will provide a more accurate prediction or measurement of the disease progression.
ACKNOWLEDGMENTS
This work was supported by NIH Grants DP3 DK110844 and UC4 DK104167.
Abbreviations:
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- AAb
autoantibody
- HbA1c
hemoglobin A1c
- FPG
fasting plasma glucose
- OGTT
oral glucose tolerance test
- ICA
islet-cell cytoplasmic autoantibody
- GAD
glutamic acid decarboxylase
- GADA
GAD autoantibody
- IA-2A
insulinoma 2- associated autoantibody
- IAA
insulin autoantibody
- ZnT8A
zinc transporter 8 autoantibody
- PTM
post-translational modifications
Footnotes
The authors declare no conflicts of interest.
All authors have read the journal’s authorship agreement and that the manuscript and approved by all named authors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Eisenbarth GS, Flier JS, Cahill G. Type-I Diabetes-Mellitus - a Chronic Autoimmune-Disease. New Engl J Med. 1986;314:1360–8. [DOI] [PubMed] [Google Scholar]
- [2].Rigby MR, Ehlers MR. Targeted immune interventions for type 1 diabetes: not as easy as it looks! Curr Opin Endocrinol 2014;21:271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ. Epidemiology of Type 1 Diabetes. Endocrinol Metab Clin North Am. 2010;39:481-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Katsarou A, Gudbjornsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, et al. Type 1 diabetes mellitus. Nat Rev Dis Primers.2017;3. [DOI] [PubMed] [Google Scholar]
- [5].Kobos E, Imiela J. Factors affecting the level of burden of caregivers of childrer with type 1 diabetes. Appl Nurs Res. 2015;28:142–9. [DOI] [PubMed] [Google Scholar]
- [6].Haugstvedt A, Wentzel-Larsen T, Rokne B, Graue M. Perceived family burden similarities and differences between mothers and fathers of children with type 1 diabets population-based study. Pediatr Diabetes. 2011;12:107–14. [DOI] [PubMed] [Google Scholar]
- [7].Zhi W, Purohit S, Carey C, Wang M, She JX. Proteomic technologies for the discovery of type 1 diabetes biomarkers. Journal of diabetes science and technology. 2010;4:993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Saisho Y, Butler AE, Manesso E, Elashoff D, Rizza RA, Butler PC. Beta-cell mass and turnover in human: effects of obesity and aging. Diabetes Care. 2013;36:111–7] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Atkinson MA, von Herrath M, Powers AC, Clare-Salzler M. Current concepts on the pathogenesis of type 1 diabetes--considerations for attempts to prevent and reverse the disease. Diabetes Care. 2015;38:979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Torn C, Mueller PW, Schlosser M, Bonifacio E, Bingley PJ, Participating L. Diabetes Antibody Standardization Program: evaluation of assays for autoantibodies to glutamic acid decarboxylase and islet antigen-2. Diabetologia. 2008;51:846–52. [DOI] [PubMed] [Google Scholar]
- [11].Schlosser M, Mueller PW, Torn C, Bonifacio E, Bingley PJ, Participating L. Diabetes Antibody Standardization Program: evaluation of assays for insulin autoantibodies. Diabetologia. 2010;53:2611–20. [DOI] [PubMed] [Google Scholar]
- [12].Schlosser M, Mueller PW, Achenbach P, Lampasona V, Bingley PJ, Participating L. Diabetes Antibody Standardization Program: First evaluation of assays for autoantibodies to IA-2beta. Diabetes Care. 2011;34:2410–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lampasona V, Schlosser M, Mueller PW, Williams AJ, Wenzlau JM, Hutton JC, et al. Diabetes antibody standardization program: first proficiency evaluation of assays for autoantibodies to zinc transporter 8. Clin Chem. 2011;57:1693–702. [DOI] [PubMed] [Google Scholar]
- [14].Sacks DB, Bruns DE, Goldstein DE, Maclaren NK, McDonald JM, Parrott M. Guidelines and recommendations for laboratory analysis in the diagnosis and management of diabetes mellitus. Clin Chem. 2002;48:436–72. [PubMed] [Google Scholar]
- [15].Ziegler AG, Rewers M, Simell O, Simell T, Lempainen J, Steck A, et al. Seroconversion to Multiple Islet Autoantibodies and Risk of Progression to Diabetes in Children. Jama-J Am Med Assoc. 2013;309:2473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Scholin A, Torn C, Nystrom L, Berne C, Arnqvist H, Blohme G, et al. Normal weight promotes remission and low number of islet antibodies prolong the duration of remission in Type 1 diabetes. Diabet Med. 2004;21:447–55. [DOI] [PubMed] [Google Scholar]
- [17].Leighton E, Sainsbury CAR, Jones GC. A Practical Review of C-Peptide Testing in Diabetes. Diabetes Ther. 2017;8:475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Szypowska A, Groele L, Wysocka-Mincewicz M, Mazur A, Lisowicz L, Ben-Skowronek I, et al. Preserved C-peptide levels in overweight or obese children with newly diagnosed type 1 diabetes. Diabetologia. 2017;60:S560–S. [Google Scholar]
- [19].Palmer JP, Fleming GA, Greenbaum CJ, Herold KC, Jansa LD, Hubert Kolb JML, 7 Kenneth S. Polonsky 8 Paolo Pozzilli 9 Jay S. Skyler 10 and Michael, et al. C-Peptide Is the Appropriate Outcome Measure for Type 1 Diabetes Clinical Trials to Preserve β-Cell Function. Diabetes. 2004;53:250–64. [DOI] [PubMed] [Google Scholar]
- [20].Knip M Disease-associated autoimmunity and prevention of insulin-dependent diabetes mellitus. Ann Med. 1997;29:447–51. [DOI] [PubMed] [Google Scholar]
- [21].Knip M, Korhonen S, Kulmala P, Veijola R, Reunanen A, Raitakari OT, et al. Prediction of type 1 diabetes in the general population. Diabetes Care. 2010;33:1206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bonifacio E Predicting type 1 diabetes using biomarkers. Diabetes Care. 2015;38:989–96. [DOI] [PubMed] [Google Scholar]
- [23].llonen J, Lempainen J, Hammais A, Laine AP, Harkonen T, Toppari J, et al. Primary islet autoantibody at initial seroconversion and autoantibodies at diagnosis of type 1 diabetes as markers of disease heterogeneity. Pediatr Diabetes. 2018;19:284–92. [DOI] [PubMed] [Google Scholar]
- [24].Endesfelder D, Hagen M, Winkler C, Haupt F, Zillmer S, Knopff A, et al. A novel approach for the analysis of longitudinal profiles reveals delayed progression to type 1 diabetes in a subgroup of multiple- islet-autoantibody-positive children. Diabetologia. 2016;59:2172–80. [DOI] [PubMed] [Google Scholar]
- [25].Vehik K, Lynch KF, Schatz DA, Akolkar B, Hagopian W, Rewers M, et al. Reversion of beta-Cell Autoimmunity Changes Risk of Type 1 Diabetes: TEDDY Study. Diabetes Care. 2016;39:1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Calderon B, Sacks DB. Islet autoantibodies and type 1 diabetes: does the evidence support screening? Clin Chem. 2014;60:438–40. [DOI] [PubMed] [Google Scholar]
- [27].Achenbach P, Koczwara K, Knopff A, Naserke H, Ziegler AG, Bonifacio E. Mature high-affinity immune responses to (pro)insulin anticipate the autoimmune cascade that leads to type 1 diabetes. J Clin Invest. 2004;114:589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bingley PJ. Clinical Applications of Diabetes Antibody Testing. J Clin Endocr Metab. 2010;95:25–33. [DOI] [PubMed] [Google Scholar]
- [29].Winter WE, Schatz DA. Autoimmune Markers in Diabetes. Clin Chem. 2011;57:168–75. [DOI] [PubMed] [Google Scholar]
- [30].Purohit S, She JX. Biomarkers for Type 1 Diabetes. Int J Clin Exp Med. 2008;1:98–116. [PMC free article] [PubMed] [Google Scholar]
- [31].Jin Y, She JX. Novel biomarkers in type 1 diabetes. The review of diabetic studies : RDS. 2012;9:224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Classification and Diagnosis of Diabetes. Diabetes Care. 2015;38:S8–S16. [DOI] [PubMed] [Google Scholar]
- [34].Cianciaruso C, Phelps EA, Pasquier M, Hamelin R, Demurtas D, Alibashe Ahmed M, et al. Primary Human and Rat beta-Cells Release the Intracellular Autoantigens GAD65, IA-2, and Proinsulin in Exosomes Together With Cytokine-Induced Enhancers of Immunity. Diabetes. 2017;66:460–73. [DOI] [PubMed] [Google Scholar]
- [35].Robitaille K, Rourke JL, McBane JE, Fu A, Baird S, Du Q, et al. High-throughput Functional Genomics Identifies Regulators of Primary Human Beta Cell Proliferation. J Biol Chem. 2016;291:4614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bader E, Migliorini A, Gegg M, Moruzzi N, Gerdes J, Roscioni SS, et al. Identification of proliferative and mature beta-cells in the islets of Langerhans. Nature. 2016;535:430–4. [DOI] [PubMed] [Google Scholar]
- [37].Rondas D, Crevecoeur I, D’Hertog W, Ferreira GB, Staes A, Garg AD, et al. Citrullinated glucose- regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes. 2015;64:573–86. [DOI] [PubMed] [Google Scholar]
- [38].El Ouaamari A, Zhou JY, Liew CW, Shirakawa J, Dirice E, Gedeon N, et al. Compensatory Islet Response to Insulin Resistance Revealed by Quantitative Proteomics. J Proteome Res. 2015;14:3111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Burch TC, Morris MA, Campbell-Thompson M, Pugliese A, Nadler JL, Nyalwidhe JO. Proteomic Analysis of Disease Stratified Human Pancreas Tissue Indicates Unique Signature of Type 1 Diabetes. PLoS One. 2015;10:e0135663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang L, Lanzoni G, Battarra M, Inverardi L, Zhang Q. Proteomic profiling of human islets collected from frozen pancreata using laser capture microdissection. J Proteomics. 2017;150:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nyalwidhe JO, Grzesik WJ, Burch TC, Semeraro ML, Waseem T, Gerling IC, et al. Comparative quantitative proteomic analysis of disease stratified laser captured microdissected human islets identifies proteins and pathways potentially related to type 1 diabetes. PLoS One. 2017;12:e0183908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhu Y, Piehowski PD, Zhao R, Chen J, Shen Y, Moore RJ, et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nature communications. 2018;9:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, Jansen E, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–94 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Klein D, Misawa R, Bravo-Egana V, Vargas N, Rosero S, Piroso J, et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. Plos One. 2013;8:e55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Qian WJ, Kaleta DT, Petritis BO, Jiang HL, Liu T, Zhang X, et al. Enhanced Detection of Low Abundance Human Plasma Proteins Using a Tandem IgY12-SuperMix Immunoaffinity Separation Strategy. Molecular & Cellular Proteomics. 2008;7:1963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tu CJ, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJC, et al. Depletion of Abundant Plasma Proteins and Limitations of Plasma Proteomics. Journal of proteome research. 2010;9:4982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jin WH, Dai J, Li SJ, Xia QC, Zou HF, Zeng R. Human plasma proteome analysis by multidimensional chromatography prefractionation and linear ion trap mass spectrometry identification. Journal of proteome research. 2005;4:613–9. [DOI] [PubMed] [Google Scholar]
- [48].Zhi WB, Sharma A, Purohit S, Miller E, Bode B, Anderson SW, et al. Discovery and Validation of Serum Protein Changes in Type 1 Diabetes Patients Using High Throughput Two Dimensional Liquid Chromatography-Mass Spectrometry and Immunoassays. Mol Cell Proteomics. 2011;10:M111.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].von Toerne C, Laimighofer M, Achenbach P, Beyerlein A, Gala TD, Krumsiek J, et al. Peptide serum markers in islet autoantibody-positive children. Diabetologia. 2017;60:287–95. [DOI] [PubMed] [Google Scholar]
- [50].Metz TO, Qian WJ, Jacobs JM, Gritsenko MA, Moore RJ, Polpitiya AD, et al. Application of proteomics in the discovery of candidate protein biomarkers in a diabetes autoantibody standardization program sample subset. Journal of proteome research. 2008;7:698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang Q, Fillmore TL, Schepmoes AA, Clauss TR, Gritsenko MA, Mueller PW, et al. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J Exp Med. 2013;210:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Moulder R, Bhosale SD, Erkkila T, Laajala E, Salmi J, Nguyen EV, et al. Serum proteomes distinguish children developing type 1 diabetes in a cohort with HLA-conferred susceptibility. Diabetes. 2015;64:2265–78. [DOI] [PubMed] [Google Scholar]
- [53].Liu CW, Bramer L, Webb-Robertson BJ, Waugh K, Rewers MJ, Zhang QB. Temporal expression profiling of plasma proteins reveals oxidative stress in early stages of Type 1 Diabetes progression. Journal of Proteomics. 2018;172:100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–50. [DOI] [PubMed] [Google Scholar]
- [55].Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–904. [DOI] [PubMed] [Google Scholar]
- [56].Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Molecular systems biology. 2017; 13:942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Eizirik DL, Cnop M. ER Stress in Pancreatic beta Cells: The Thin Red Line Between Adaptation and Failure. Sci Signaling. 2010;3:pe7. [DOI] [PubMed] [Google Scholar]
- [58].Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Massa O, Alessio M, Russo L, Nardo G, Bonetto V, Bertuzzi F, et al. Serological Proteome Analysis (SERPA) as a tool for the identification of new candidate autoantigens in type 1 diabetes. Journal of Proteomics. 2013;82:263–73. [DOI] [PubMed] [Google Scholar]
- [60].McGinty JW, Chow IT, Greenbaum C, Odegard J, Kwok WW, James EA. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes. 2014;63:3033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yu WL, Sun Y. Citrullinated Glucose-Regulated Protein 78 Is an Autoantigen in Type 1 Diabetes. Diabetes 2015;64: 573–586. Diabetes. 2015;64:E4-E. [DOI] [PubMed] [Google Scholar]
- [62].van Lummel M, Duinkerken G, van Veelen PA, de Ru A, Cordfunke R, Zaldumbide A, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes. 2014;63:237–47. [DOI] [PubMed] [Google Scholar]
- [63].Strollo R, Rizzo P, Spoletini M, Landy R, Hughes C, Ponchel F, et al. HLA-dependent autoantibodies against post-translationally modified collagen type II in type 1 diabetes mellitus. Diabetologia. 2013;56:563–72. [DOI] [PubMed] [Google Scholar]
- [64].Strollo R, Vinci C, Napoli N, Pozzilli P, Ludvigsson J, Nissim A. Antibodies to post-translationally modified insulin as a novel biomarker for prediction of type 1 diabetes in children. Diabetologia. 2017;60:1467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Doran TM, Morimoto J, Simanski S, Koesema EJ, Clark LF, Pels K, et al. Discovery of Phosphorylated Peripherin as a Major Humoral Autoantigen in Type 1 Diabetes Mellitus. Cell chemical biology. 2016;23:618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mannering SI, Harrison LC, Williamson NA, Morris JS, Thearle DJ, Jensen KP, et al. The insulin A- chain epitope recognized by human T cells is posttranslationally modified. J Exp Med. 2005;202:1191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bezu L, Sauvat A, Humeau J, Gomes-da-Silva LC, Iribarren K, Forveille S, et al. eIF2alpha phosphorylation is pathognomonic for immunogenic cell death. Cell Death Differ. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ganesan L, Levental I. Pharmacological Inhibition of Protein Lipidation. J Membr Biol. 2015;248:929–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Freitas PAC, Ehlert LR, Camargo JL. Glycated albumin: a potential biomarker in diabetes. Arch Endocrinol Metab. 2017;61:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nature Reviews Endocrinology. 2009;5:219–26. [DOI] [PubMed] [Google Scholar]
- [72].Stechova K, Bohmova K, Vrabelova Z, Sepa A, Stadlerova G, Zacharovova K, et al. High T-helper-1 cytokines but low T-helper-3 cytokines, inflammatory cytokines and chemokines in children with high risk of developing type 1 diabetes. Diabetes Metab Res Rev. 2007;23:462–71. [DOI] [PubMed] [Google Scholar]
- [73].Yeung WCG, Al-Shabeeb A, Pang CNI, Wilkins MR, Catteau J, Howard NJ, et al. Children With Islet Autoimmunity and Enterovirus Infection Demonstrate a Distinct Cytokine Profile. Diabetes. 2012;61:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Takahashi K, Ohara M, Sasai T, Homma H, Nagasawa K, Takahashi T, et al. Serum CXCL1 concentrations are elevated in type 1 diabetes mellitus, possibly reflecting activity of anti-islet autoimmune activity. Diabetes Metab Res Rev 2011;27:830–3. [DOI] [PubMed] [Google Scholar]
- [75].Purohit S, Sharma A, Hopkins D, Steed L, Bode B, Anderson SW, et al. Large-Scale Discovery and Validation Studies Demonstrate Significant Reductions in Circulating Levels of IL8, IL-1Ra, MCP-1, and MIP-1 beta in Patients With Type 1 Diabetes. J Clin Endocrinol Metab. 2015;100:E1179–E87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mack CL. Serum cytokines as biomarkers of disease and clues to pathogenesis. Hepatology. 2007;46:6–8. [DOI] [PubMed] [Google Scholar]
- [77].Ali AM, DiPersio JF, Schroeder MA. The Role of Biomarkers in the Diagnosis and Risk Stratification of Acute Graft-versus-Host Disease: A Systematic Review. Biol Blood Marrow Transplant. 2016;22:1552–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1 alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci U S A. 2011;108:8885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Roder ME, Knip M, Hartling SG, Karjalainen J, Akerblom HK, Binder C, et al. Disproportionately Elevated Proinsulin Levels Precede the Onset of Insulin-Dependent Diabetes-Mellitus in Siblings with Low First-Phase Insulin Responses. J Clin Endocrinol Metab. 1994;79:1570–5. [DOI] [PubMed] [Google Scholar]
- [80].Sims EK, Chaudhry Z, Watkins R, Syed F, Blum J, Ouyang FQ, et al. Elevations in the Fasting Serum Proinsulin-to-C-Peptide Ratio Precede the Onset of Type 1 Diabetes. Diabetes Care. 2016;39:1519–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Truyen I, Pauw P, Jorgensen PN, Van Schravendijk C, Ubani O, Decochez K, et al. Proinsulin levels and the proinsulin : C-peptide ratio complement autoantibody measurement for predicting type 1 diabetes. Diabetologia. 2005;48:2322–9. [DOI] [PubMed] [Google Scholar]
- [82].McLaughlin KA, Richardson CC, Ravishankar A, Brigatti C, Liberati D, Lampasona V, et al. Identification of Tetraspanin-7 as a Target of Autoantibodies in Type 1 Diabetes. Diabetes. 2016;65:1690–8. [DOI] [PubMed] [Google Scholar]
- [83].Van De Winkel M, Smets G, Gepts W, Pipeleers D. Islet cell surface antibodies from insulin- dependent diabetics bind specifically to pancreatic B cells. J Clin Invest. 1982;70:41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Astorri E, Guglielmi C, Bombardieri M, Alessandri C, Buzzetti R, Maggi D, et al. Circulating Reg1alpha proteins and autoantibodies to Reg1alpha proteins as biomarkers of beta-cell regeneration and damage in type 1 diabetes. Horm Metab Res. 2010;42:955–60. [DOI] [PubMed] [Google Scholar]
- [85].Li SW, Koya V, Li Y, Donelan W, Lin P, Reeves WH, et al. Pancreatic duodenal homeobox 1 protein is a novel beta-cell-specific autoantigen for type I diabetes. Lab Invest. 2010;90:31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Shehadeh N, Pollack S, Wildbaum G, Zohar Y, Shafat I, Makhoul R, et al. Selective autoantibody production against CCL3 is associated with human type 1 diabetes mellitus and serves as a novel biomarker for its diagnosis. J Immunol. 2009;182:8104–9. [DOI] [PubMed] [Google Scholar]
- [87].Fierabracci A, Saura F. Identification of a common autoantigenic epitope of protein disulfide isomerase, golgin-160 and voltage-gated potassium channel in type 1 diabetes. Diabetes Res Clin Pract. 2010;88:e14–6. [DOI] [PubMed] [Google Scholar]
- [88].Endo T, Takizawa S, Tanaka S, Takahashi M, Fujii H, Kamisawa T, et al. Amylase alpha-2A autoantibodies: novel marker of autoimmune pancreatitis and fulminant type 1 diabetes. Diabetes. 2009;58:732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wan EC, Gordon TP, Jackson MW. Autoantibodies to calcium channels in type 1 diabetes mediate autonomic dysfunction by different mechanisms in colon and bladder and are neutralized by antiidiotypic antibodies. J Autoimmun. 2008;31:66–72. [DOI] [PubMed] [Google Scholar]
- [90].Jackson MW, Gordon TP. A novel impedance-based cellular assay for the detection of anti-calcium channel autoantibodies in type 1 diabetes. J Immunol Methods. 2010;361:31–6. [DOI] [PubMed] [Google Scholar]
- [91].Park SG, Park HS, Jeong IK, Cho YM, Lee HK, Kang YS, et al. Autoantibodies against aminoacyl-tRNA synthetase: novel diagnostic marker for type 1 diabetes mellitus. Biomarkers. 2010;15:358–66. [DOI] [PubMed] [Google Scholar]
- [92].Kaufman DL, Erlander MG, Clare-Salzler M, Atkinson MA, Maclaren NK, Tobin AJ. Autoimmunity to two forms of glutamate decarboxylase in insulin-dependent diabetes mellitus. J Clin Invest. 1992;89:283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wasmeier C, Hutton JC. Molecular cloning of phogrin, a protein-tyrosine phosphatase homologue localized to insulin secretory granule membranes. The Journal of biological chemistry. 1996;271:18161–70. [DOI] [PubMed] [Google Scholar]
- [94].Liu H, Wang Z, Li S, Zhang Y, Yan YC, Li YP. Utilization of an intron located polyadenlyation site resulted in four novel glutamate decarboxylase transcripts. Mol Biol Rep. 2009;36:1469–74. [DOI] [PubMed] [Google Scholar]
- [95].Diez J, Park Y, Zeller M, Brown D, Garza D, Ricordi C, et al. Differential splicing of the IA-2 mRNA in pancreas and lymphoid organs as a permissive genetic mechanism for autoimmunity against the IA-2 type 1 diabetes autoantigen. Diabetes. 2001;50:895–900. [DOI] [PubMed] [Google Scholar]
- [96].Marchand L, Polychronakos C. Evaluation of polymorphic splicing in the mechanism of the association of the insulin gene with diabetes. Diabetes. 2007;56:709–13. [DOI] [PubMed] [Google Scholar]
- [97].Zavialov A, Ankelo M, Westerlund-Karlsson A, Knip M, Ilonen J, Hinkkanen A. Novel fusion proteins in the analysis of diabetes-associated autoantibodies to GAD65 and IA-2. J Immunol Methods. 2000;246:91–6. [DOI] [PubMed] [Google Scholar]
- [98].Massa O, Alessio M, Russo L, Nardo G, Bonetto V, Bertuzzi F, et al. Serological Proteome Analysis (SERPA) as a tool for the identification of new candidate autoantigens in type 1 diabetes. J Proteomics. 2013;82:263–73. [DOI] [PubMed] [Google Scholar]
- [99].Wenzlau JM, Hutton JC. Novel diabetes autoantibodies and prediction of type 1 diabetes. Curr Diab Rep 2013;13:608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem. 2007;53:2215. [DOI] [PubMed] [Google Scholar]
- [101].Wang W, Kong P, Ma G, Li L, Zhu J, Xia T, et al. Characterization of the release and biological significance of cell-free DNA from breast cancer cell lines. Oncotarget. 2017;8:43180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Syed F, Evans-Molina C. Nucleic acid biomarkers of beta cell stress and death in type 1 diabetes. Curr Opin Endocrinol. 2016;23:312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Herold KC, Usmani-Brown S, Ghazi T, Lebastchi J, Beam CA, Bellin MD, et al. beta Cell death and dysfunction during type 1 diabetes development in at-risk individuals. J Clin Invest. 2015;125:1163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V, et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. P Natl Acad Sci USA. 2011;108:19018–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113:E1826–E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Usmani-Brown S, Lebastchi J, Steck AK, Beam C, Herold KC, Ledizet M. Analysis of beta-cell death in type 1 diabetes by droplet digital PCR. Endocrinology. 2014;155:3694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fisher MM, Watkins RA, Blum J, Evans-Molina C, Chalasani N, DiMeglio LA, et al. Elevations in Circulating Methylated and Unmethylated Preproinsulin DNA in New-Onset Type 1 Diabetes. Diabetes. 2015;64:3867–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lebastchi J, Deng SY, Lebastchi AH, Beshar I, Gitelman S, Willi S, et al. Immune Therapy and betaCell Death in Type 1 Diabetes. Diabetes. 2013;62:1676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Olsen JA, Kenna LA, Spelios MG, Hessner MJ, Akirav EM. Circulating Differentially Methylated Amylin DNA as a Biomarker of beta- Cell Loss in Type 1 Diabetes. Plos One. 2016;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kallionpaa H, Elo LL, Laajala E, Mykkanen J, Ricano-Ponce I, Vaarma M, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63:2402–14. [DOI] [PubMed] [Google Scholar]
- [111].Jin YL, Sharma A, Bai S, Davis C, Liu H, Hopkins D, et al. Risk of Type 1 Diabetes Progression in Islet Autoantibody-Positive Children Can Be Further Stratified Using Expression Patterns of Multiple Genes Implicated in Peripheral Blood Lymphocyte Activation and Function. Diabetes. 2014;63:2506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ferreira RC, Guo H, Coulson RMR, Smyth DJ, Pekalski ML, Burren OS, et al. A Type I Interferon Transcriptional Signature Precedes Autoimmunity in Children Genetically at Risk for Type 1 Diabetes. Diabetes. 2014;63:2538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Irvine KM, Gallego P, An XY, Best SE, Thomas G, Wells C, et al. Peripheral Blood Monocyte Gene Expression Profile Clinically Stratifies Patients With Recent-Onset Type 1 Diabetes. Diabetes. 2012;61:1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Assmann TS, Recamonde-Mendoza M, De Souza BM, Crispim D. MicroRNA expression profiles and type 1 diabetes mellitus: systematic review and bioinformatic analysis. Endocr Connect. 2017;6:773–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Seyhan AA, Lopez YON, Xie H, Yi FC, Mathews C, Pasarica M, et al. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Scientific reports. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SLT, Wong MTK, et al. Circulating miRNA Profiles in Patients with Metabolic Syndrome. J Clin Endocrinol Metab. 2012;97:E2271–E6. [DOI] [PubMed] [Google Scholar]
- [117].Latreille M, Herrmanns K, Renwick N, Tuschl T, Malecki MT, McCarthy MI, et al. miR-375 gene dosage in pancreatic beta-cells: implications for regulation of beta-cell mass and biomarker development. J Mol Med (Berl). 2015;93:1159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Marchand L, Jalabert A, Meugnier E, Van den Hende K, Fabien N, Nicolino M, et al. miRNA-375 a Sensor of Glucotoxicity Is Altered in the Serum of Children with Newly Diagnosed Type 1 Diabetes. Journal of Diabetes Research. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Erener S, Marwaha A, Tan R, Kieffer TJ. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. Diabetologia. 2016;59:S161–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Osipova J, Fischer DC, Dangwal S, Volkmann I, Widera C, Schwarz K, et al. Diabetes-Associated MicroRNAs in Pediatric Patients With Type 1 Diabetes Mellitus: A Cross-Sectional Cohort Study. J Clin Endocrinol Metab. 2014;99:E1661–E5. [DOI] [PubMed] [Google Scholar]
- [121].Nielsen LB, Wang C, Sorensen K, Bang-Berthelsen CH, Hansen L, Andersen MLM, et al. Circulating Levels of MicroRNA from Children with Newly Diagnosed Type 1 Diabetes and Healthy Controls: Evidence That miR-25 Associates to Residual Beta-Cell Function and Glycaemic Control during Disease Progression (vol 2012, 896362, 2012). Experimental Diabetes Research. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a Biomarker of betaCell Death and Diabetes in Mice. Endocrinology. 2013;154:603–8. [DOI] [PubMed] [Google Scholar]
- [123].Nabih ES, Andrawes NG. The Association Between Circulating Levels of miRNA-181a and Pancreatic Beta Cells Dysfunction via SMAD7 in Type 1 Diabetic Children and Adolescents. J Clin Lab Anal 2016;30:727–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–U72. [DOI] [PubMed] [Google Scholar]
- [125].Lakhter AJ, Pratt RE, Moore RE, Doucette KK, Maier BF, DiMeglio LA, et al. Beta cell extracellular vesicle miR-21–5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Garcia-Contreras M, Shah SH, Tamayo A, Robbins PD, Golberg RB, Mendez AJ, et al. Plasma- derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Scientific reports. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Pflueger M, Seppanen-Laakso T, Suortti T, Hyotylainen T, Achenbach P, Bonifacio E, et al. Age- and Islet Autoimmunity-Associated Differences in Amino Acid and Lipid Metabolites in Children at Risk for Type 1 Diabetes. Diabetes. 2011;60:2740–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Bervoets L, Massa G, Guedens W, Louis E, Noben JP, Adriaensens P. Metabolic profiling of type 1 diabetes mellitus in children and adolescents: a case-control study. Diabetol Metab Syndr. 2017;9:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Leslie RD, Beyan H. Metabolomics makes a mark: early changes associated with autoimmune diabetes. Diabetes. 2011;60:2688–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Fox TE, Bewley MC, Unrath KA, Pedersen MM, Anderson RE, Jung DY, et al. Circulating sphingolipid biomarkers in models of type 1 diabetes. J Lipid Res. 2011;52:509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].La Torre D, Seppanen-Laakso T, Larsson HE, Hyotylainen T, Ivarsson SA, Lernmark A, et al. Decreased cord-blood phospholipids in young age-at-onset type 1 diabetes. Diabetes. 2013;62:3951–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Oresic M, Simell S, Sysi-Aho M, Nanto-Salonen K, Seppanen-Laakso T, Parikka V, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Oresic M, Gopalacharyulu P, Mykkanen J, Lietzen N, Makinen M, Nygren H, et al. Cord serum lipidome in prediction of islet autoimmunity and type 1 diabetes. Diabetes. 2013;62:3268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Shi T, Song E, Nie S, Rodland KD, Liu T, Qian WJ, et al. Advances in targeted proteomics and applications to biomedical research. Proteomics. 2016;16:2160–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Garcia-Contreras M, Brooks RW, Boccuzzi L, Robbins PD, Ricordi C. Exosomes as biomarkers and therapeutic tools for type 1 diabetes mellitus. Eur Rev Med Pharmacol Sci. 2017;21:2940–56. [PubMed] [Google Scholar]
- [136].Saeedi Borujeni MJ, Esfandiary E, Taheripak G, Codoner-Franch P, Alonso-Iglesias E, Mirzaei H. Molecular aspects of diabetes mellitus: Resistin, microRNA, and exosome. J Cell Biochem. 2018;119:1257–72. [DOI] [PubMed] [Google Scholar]
- [137].Tramontano AF, Lyubarova R, Tsiakos J, Palaia T, Deleon JR, Ragolia L. Circulating endothelial microparticles in diabetes mellitus. Mediators Inflamm. 2010;2010:250476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lietz CB, Toneff T, Mosier C, Podvin S, O’Donoghue AJ, Hook V. Phosphopeptidomics Reveals Differential Phosphorylation States and Novel SxE Phosphosite Motifs of Neuropeptides in Dense Core Secretory Vesicles. J Am Soc Mass Spectr. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Berezin AE, Kremzer AA, Samura TA, Berezina TA, Kruzliak P. Impaired immune phenotype of circulating endothelial-derived microparticles in patients with metabolic syndrome and diabetes mellitus. J Endocrinol Invest. 2015;38:865–74. [DOI] [PubMed] [Google Scholar]


