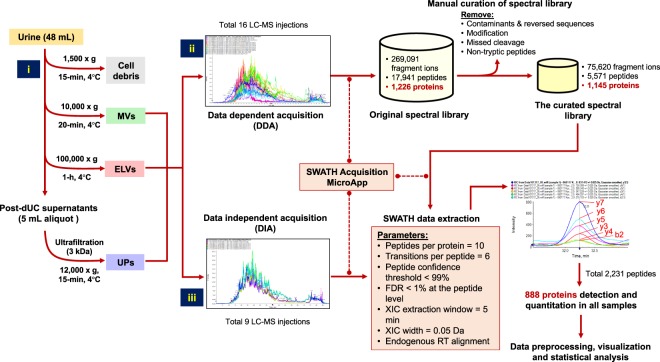
Figure 1.
The schematic diagram represents the entire process of ELV-SWATH-MS workflow. Three main steps included; (i) the isolation of urinary extracellular vesicles including exosome-like vesicles (ELVs) and microvesicles (MVs) by differential ultracentrifugation and subsequently concentrating urine proteins (UPs) by ultrafiltration (3-kDa cutoff); (ii) the spectral library generation based on all identified fragment ions, peptides and proteins from data-dependent acquisition (DDA) of 16 protein fractionations of ELVs, MVs and UPs and a manual curation to remove the sources of variability in targeted proteomic analysis; (iii) SWATH analysis using predefined targets in the curated spectral library for targeted data extraction and peptide quantitation from the digital protein records of ELVs, MVs and UPs generated by data-independent acquisition (DIA).