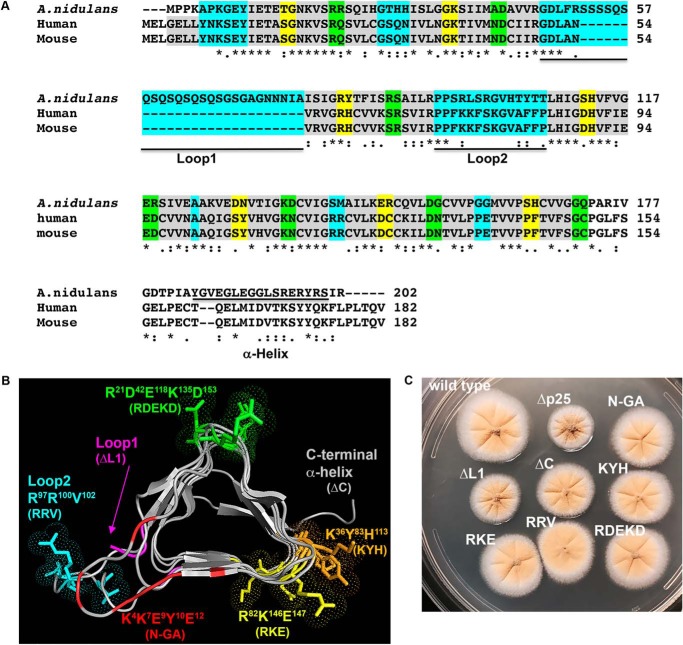
Figure 3.
Analysis of A. nidulans p25 structure and colony phenotypes of various p25 mutants. A, sequence alignment of p25. The first four amino acids of the β-strands are highlighted in gray (each β-strand contains six amino acids with the last two form the turn). Three turns in between strands are colored as yellow, green, and cyan, respectively. The two loops (loop1 and loop2) and the C-terminal α-helix (α-helix) are underlined. The alignment was done using CLUSTALW. Residues that are identical (*), strongly similar (:) or weakly similar (.) are indicated below the sequences. B, model of A. nidulans p25 and positions of the amino acids mutated in this study. The names of the corresponding mutants are labeled in the same color below the names of the regions or the amino acid clusters (each of them is highlighted using the same color as that of the label). The β-strands of the LβH as well as the C-terminal α-helix (labeled on top) are highlighted in gray, and the position of loop1 is indicated with an arrow. C, colony phenotypes of a WT control strain containing p25–GFP (wild type) and various p25 mutant strains.