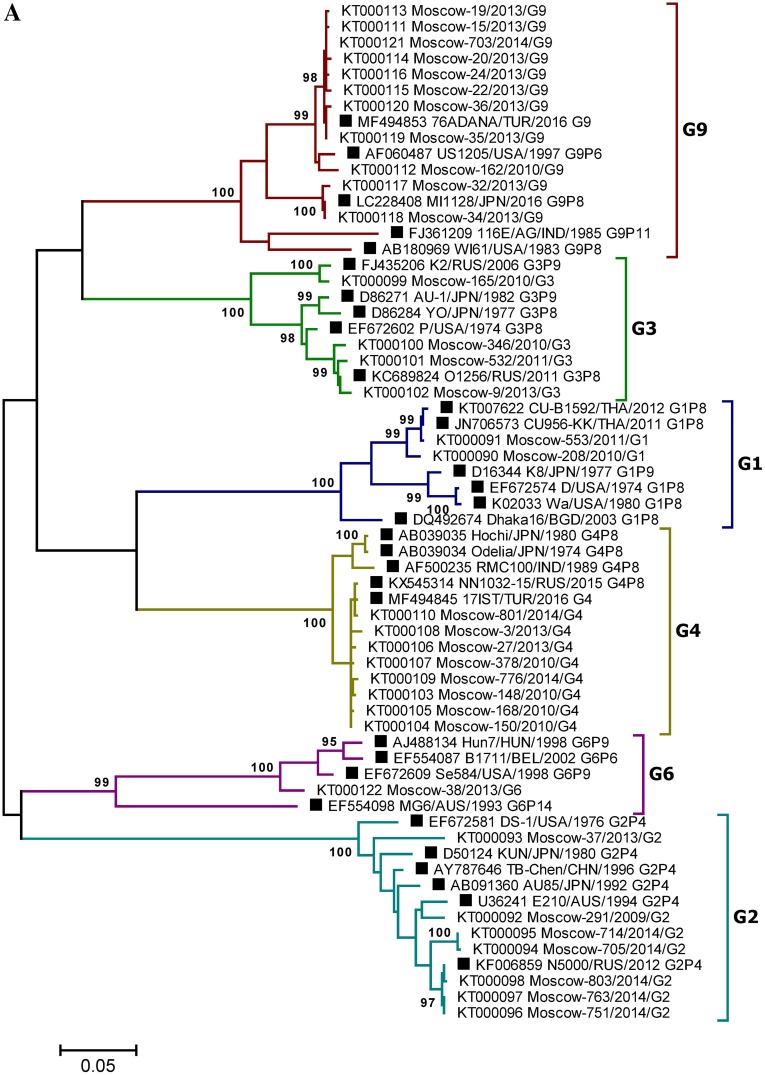
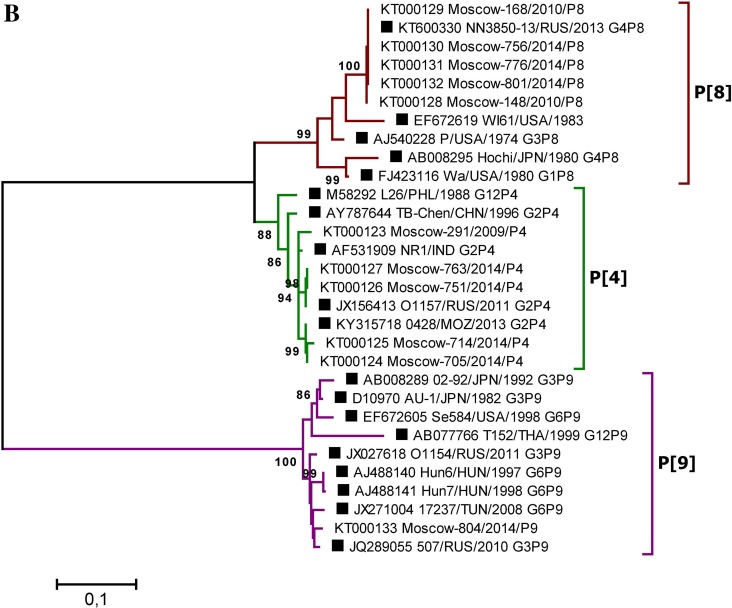
Fig. 4.
A Phylogenetic tree based on partial nucleotide sequences of the RVA VP7 (A) and VP4 (B) gene from sequenced clinical samples and reference strains. Phylogenetic trees were generated using MEGA6 (Tamura et al. 2013) with the maximum likelihood method and the Kimura two-parameter model, based on the partial nucleotide sequences of the RVA VP7 and VP4 genes. The names of clinical samples include the GenBank accession number, the sample isolation location (Moscow), a laboratory identification number, the year of isolation, and the gene variant determined by PCR. Reference strain names are indicated by GenBank accession number, strain (isolate) name, country and year of isolation, and genotype. Three-letter country codes were used according to ISO 3166-1. Numbers at the nodes are bootstrap values based on 1000 replications. Reference strains are labeled as filled square. Bootstrap cut-off values are 95% (A) and 85% (B).