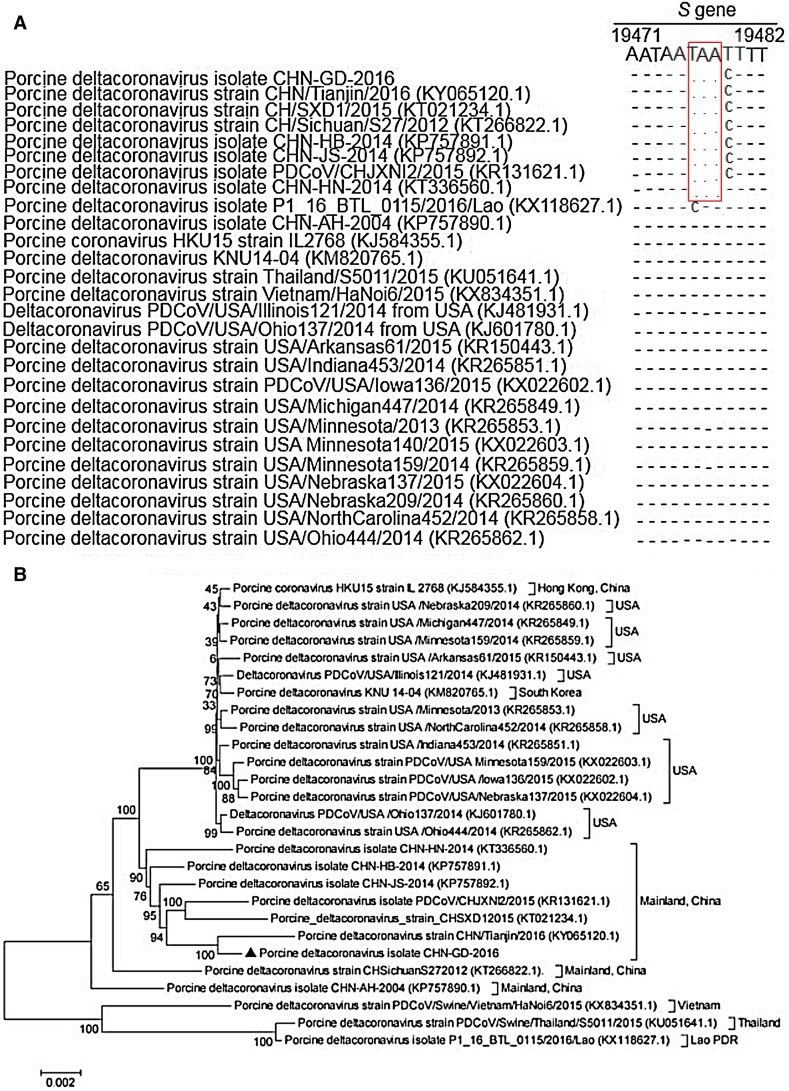
Fig. 4.
Sequence alignments partial spike genes of different porcine deltacoronavirus strains and phylogenetic tree constructed on the basis of the whole-genome nucleotide sequences of 27 PDCoV strains from different countries. A A multiple sequence of partial spike (S) genes was constructed with Clustal W in the DNAStar software. Red rectangles indicate the 3-nt deletion sites. B The dendrogram was constructed using the neighbor-joining method in the MEGA software package, version 5 (http://www.megasoftware.net). Bootstrap resampling (1000 replication) was performed, and bootstrap values are indicated for each node. Reference sequence obtained from GenBank are indicated by strain name. The scale bar represents 0.002 nucleotide substitutions per site.