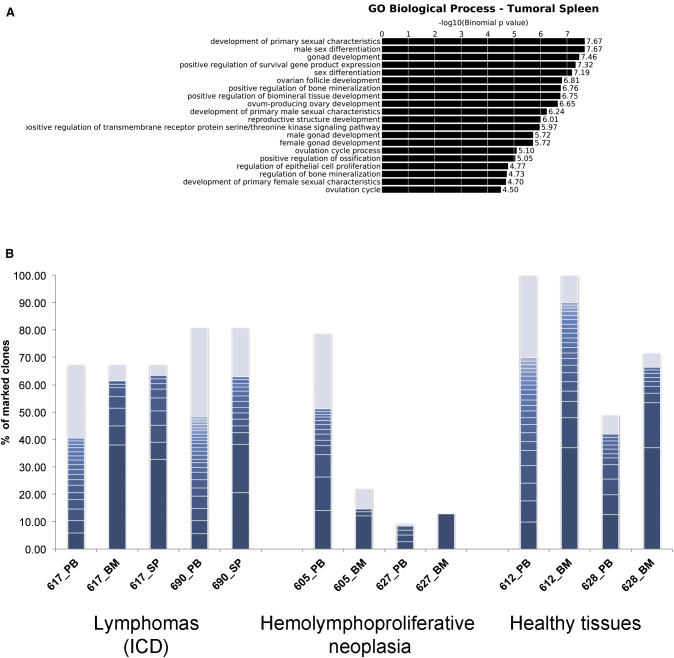
Figure 4.
Gene Ontology Analysis and Abundance Analysis of ISs from Infiltrated Tumors
DNA extracted from infiltrated tumor spleens was analyzed for vector integration (mice 617 and 690). (A) GO analysis was performed using the GREAT software. The integrations retrieved from tumors showed overrepresentations of the gene functions indicated on the left. Only gene classes with a false discovery rate of less than 0.05 were considered. We interrogated the Biological Process database. (B) Histograms of the percentage of sequence reads (y axis) for unequivocally mapped ISs from mice that developed lymphomas and hemolymphoproliferative neoplasia and healthy controls. The number of reads for each IS was normalized to the total number of sequence reads from the same time point and for the engraftment level. ISs below 1% were collapsed and are shown in gray.