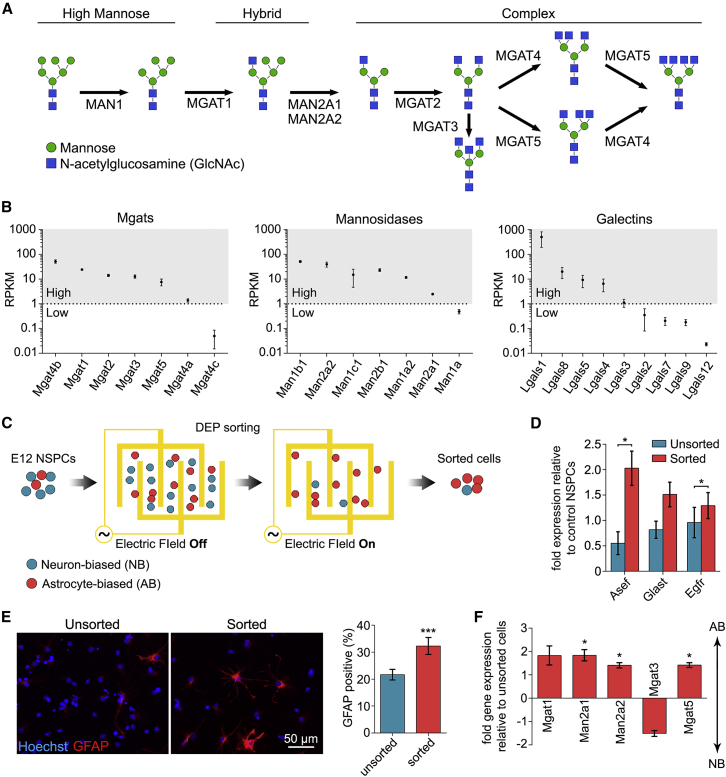
Figure 2.
N-Glycan Branching Correlates with NSPC Fate
(A) Schematic representation of the N-glycans formed by glycosylation enzymes, culminating in the formation of highly branched, complex N-glycans.
(B) RNA sequencing analysis of E12 NSPCs. Average RPKM (reads per kilobase of transcript per million mapped reads) values are organized from high (>1 RPKM) to low (<1 RPKM) expression.
(C) In DEP sorting, E12 NSPCs are randomly distributed when the electric field is off. When the electric field is on, astrocyte-biased cells are attracted to electrode edges and neuron-biased cells are removed by flow, leaving an astrocyte-biased population of sorted cells.
(D) qRT-PCR analysis of astrocyte progenitor marker expression indicates that sorted cells have significantly increased Asef (p = 0.0239) and Egfr (p = 0.0273) expression (paired Student's t test).
(E) Sorted cells generate more GFAP-positive cells after differentiation (p = 0.0002, paired Student's t test).
(F) qRT-PCR analysis shows that sorted cells express higher levels of branching enzymes (Man2a1, p = 0.0433; Man2a2, p = 0.0474; Mgat5, p = 0.0314, paired Student's t test sorted versus unsorted), while Mgat3 that prevents branching appears decreased.
All error bars represent SEM. N = 3 or more independent biological repeats (∗p < 0.05, ∗∗∗p < 0.001).