Figure 4.
Comparison of HIT-Cas9 with Existing Drug-Inducible Designs
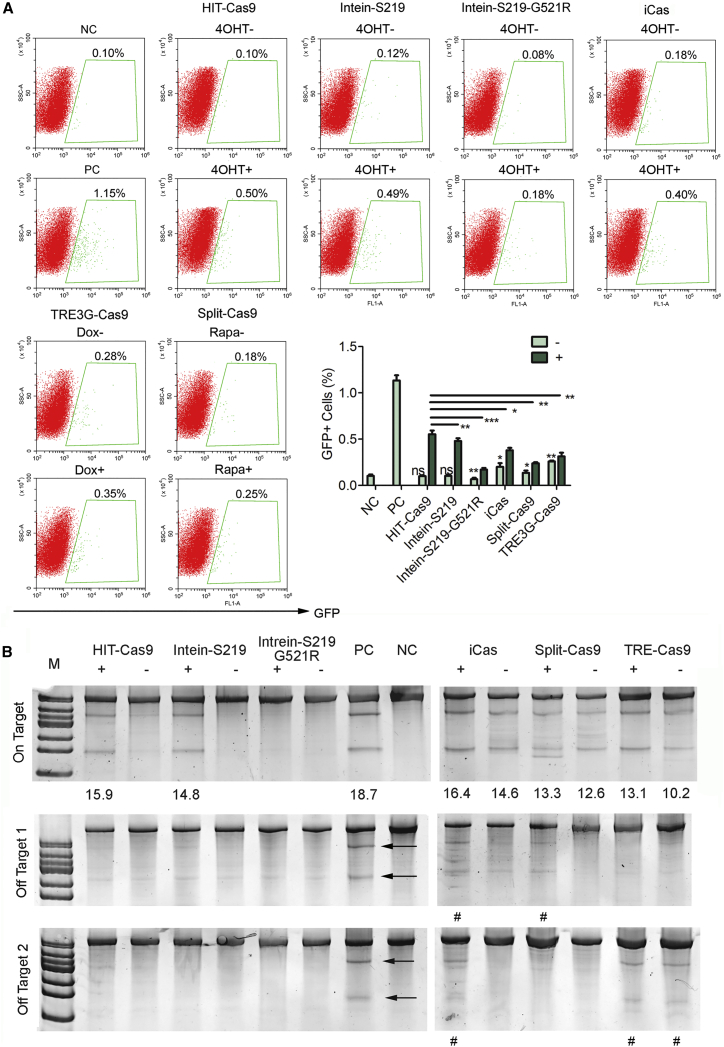
(A) Comparative analyses of drug-inducible efficiency and background activity of HIT-Cas9 and previously reported designs using the FCR assay. Representative plots (top) and quantifications (right bottom) were shown. NC, cells transfected with an unrelated sgRNA; PC, cells transfected with Cas9-NLS and BFP sgRNA. Data showed mean ± SD. n = 3 biological replicates. ns, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001; Student’s t test. (B) Drug-inducible genome editing of an EMX1 site and two off-target sites examined in Surveyor assay. Percentages (%) of indel, if detected, were listed at the bottom of each lane for on-target site analyses. Arrows point to off-target cleavage bands from NLS-Cas9. # denotes off-target activities for existing designs.