Fig. 10.
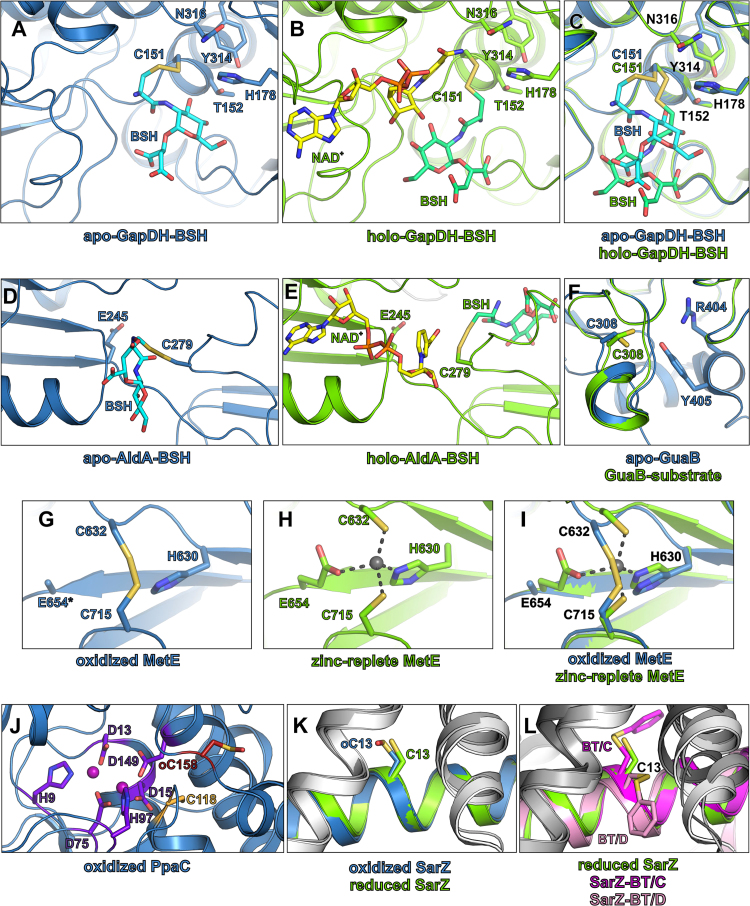
Structural insights into conserved targets for S-bacillithiolation. (A, B) The S-bacillithiolated active site pocket of S. aureus GapDH in the apo- (A) and holo-enzyme (B). (C) The superposition of the GapDH active site of the apo- (blue) and holo-enzyme (green). The molecular docking of BSH was performed on the GapDH apo- (PDB ID: 3LC7) and holo-enzyme structures (PDB ID: 3LVF) (D, E) The S-bacillithiolated active site pocket of S. aureus AldA in the apo- (blue) (D) and holo-enzyme (green) (E). Molecular docking of BSH was performed on the AldA structure (PDB ID: 3TY7). (F) The active site of the B. anthracis GuaB in the apo-enzyme (blue) (PDB ID: 3TSB) and substrate-bound structure (green) (PDB ID: 3USB), the substrate is not shown. (G, H) The active site of the S. mutans MetE in the oxidized (PDB ID: 3L7R) (G) and in the zinc-replete structure (PDB ID: 3T0C) (H). In the oxidized structure, C632 and C715 form an intramolecular disulfide bond. The asterisk indicates a glutamate residue that was modelled as alanine due to the lack of the electron density at this position. (I) The superposition of the active site of the MetE oxidized (blue) and zinc-replete (green) structures. The zinc cation is shown as gray sphere. (J) The active site of the S. aureus PpaC (PDB ID: 4RPA). The active site residues and the neighboring cysteine residues are shown as sticks and coloured by atom type. The colour of the carbon atoms allows to distinguish between active site residues (purple) and neighboring cysteine residues that were found S-bacillithiolated (dark red) or were not S-bacillithiolated (light orange). The manganese cations are shown as pink spheres. (K, L) The redox-active cysteine residue of S. aureus SarZ. (K) Superposition of the reduced (blue) (PDB ID: 3HSE) and oxidized form (green) (PDB ID: 3HRM) of SarZ. (L) Superposition of the reduced (blue) and benzene thiol-bound SarZ in chain C (magenta) and in chain D (light pink) (PDB ID: 3HSR). One monomer of the SarZ dimer is shown in blue (reduced SarZ), green (oxidized SarZ), magenta (benzene thiol-bound SarZ chain C) or light pink (benzene thiol-bound SarZ chain D), whereas the other monomer is shown in shades of gray. All the important residues in all panels are shown as sticks and coloured by atom types (oxygen-red, nitrogen-blue, sulfur-yellow, phosphor-orange, carbon-variable) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).