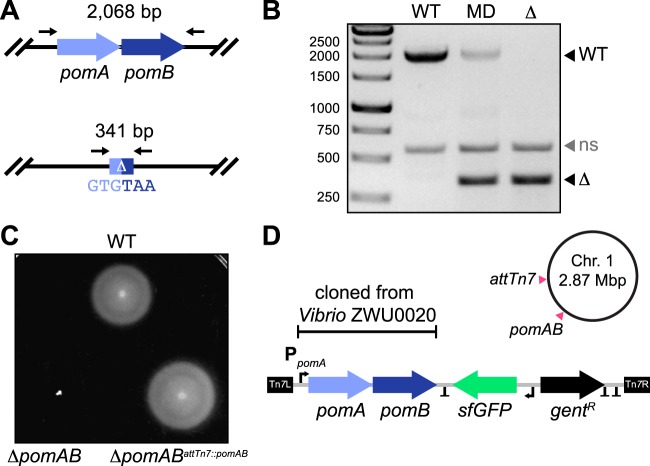
FIG 5.
Gene deletion and complementation with modernized engineering vectors. (A) (Top) Wild-type pomAB locus in Vibrio ZWU0020. (Bottom) Result of markerless pomAB deletion via allelic exchange. Black arrows mark approximate primer annealing sites for genotyping, and the size of each amplification product is indicated. (B) Agarose gel showing PCR-based genotyping of the wild-type strain (WT), a merodiploid strain (MD), and a ΔpomAB (Δ) mutant. Migration distances of WT and mutant alleles are indicated. ns, nonspecific amplification product. (C) Swim motility of the WT strain, the ΔpomAB mutant, and the complemented ΔpomABattTn7::pomAB variant in 0.2% tryptic soy agar at 30°C. (D) Shown is a schematic of the Tn7 transposon from pTn7xTS-sfGFP used for complementation, which was modified to carry the native pomAB locus of Vibrio ZWU0020. Also depicted are the relative positions where the pomAB genes were deleted and reintroduced at the Tn7 insertion site (attTn7) on chromosome 1 of Vibrio ZWU0020. T, transcriptional terminators; Tn7L and Tn7R, Tn7 inverted repeats; PpomA, native pomA promoter; gentR, gentamicin resistance gene; sfGFP, fluorescent tag.