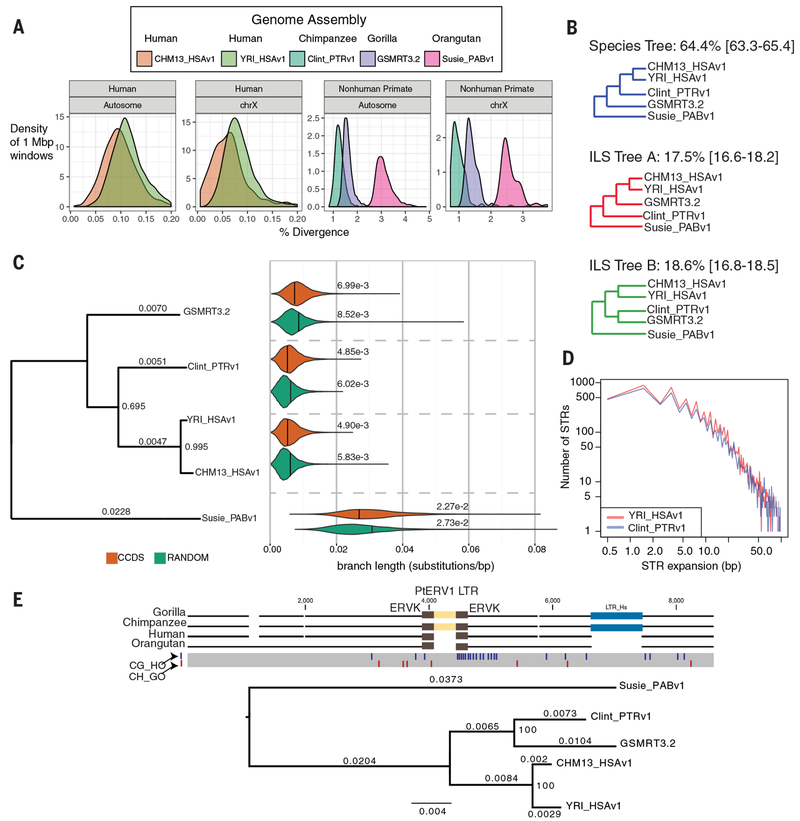
Fig. 2. Ape genetic diversity and lineage sorting.
a) Single-nucleotide variant (SNV) divergence between each primate assembly and GRCh38 was calculated in 1 Mbp non-overlapping windows across all autosomes and chromosome X (excluding X-Y homologous regions). Mean autosomal divergence is 1.27+/−0.20% (human-chimpanzee), 1.61+/−0.21% (human-gorilla) and 3.12+/−0.33% (human-orangutan). The African genome (YRI_HSAv1) shows a 17% increase in SNV diversity. b) Proportion of phylogenetic trees supporting standard species topology and incomplete lineage sorting (ILS). The mean and 95% confidence intervals are based on 100 genome-wide permutations. c) A phylogenetic tree (maximum clade credibility consensus tree) comparing genic regions (~9,000 consensus CDS (CCDS) and 1,000 bp flanking sequence [orange]) to a randomly genome-shuffled set matched to CDS lengths (green). The analysis excludes regions of SDs, SVs and large tandem repeats. Branch lengths (above the lines) and proportion of trees supporting each bifurcation (internal nodes) are shown. Violin plots summarize the distribution and mean divergence (substitutions/bp) for a subset of trees consistent with the species tree. YRI_HSAv1 is the representative human in the violin plots. d) A comparison of the expanded STR sequences (n = 16,138 loci) between human (African) and chimpanzee ab initio genome assemblies shows little to no species bias (0.02 bp). e) A multiple sequence alignment (MSA) of ape genomes (gorilla BAC CH277-16N20, chimpanzee CH251-550G17) identifies an orthologous 379 bp PtERV1 element nested within another LTR and shared between gorilla and chimpanzee. A maximum likelihood phylogenetic tree (GTR+Gamma) built from 12,108 bp supporting ILS. Single-nucleotide polymorphisms that support chimpanzee-gorilla sorting (CG_HO) are shown as blue lines and the red lines show single-nucleotide polymorphisms supporting the species tree (CH_GO). Branch lengths (substitutions per site) are shown above the lineages and internal nodes are labeled with bootstrap support (proportion of replicates supporting split; 1,000 replicates).