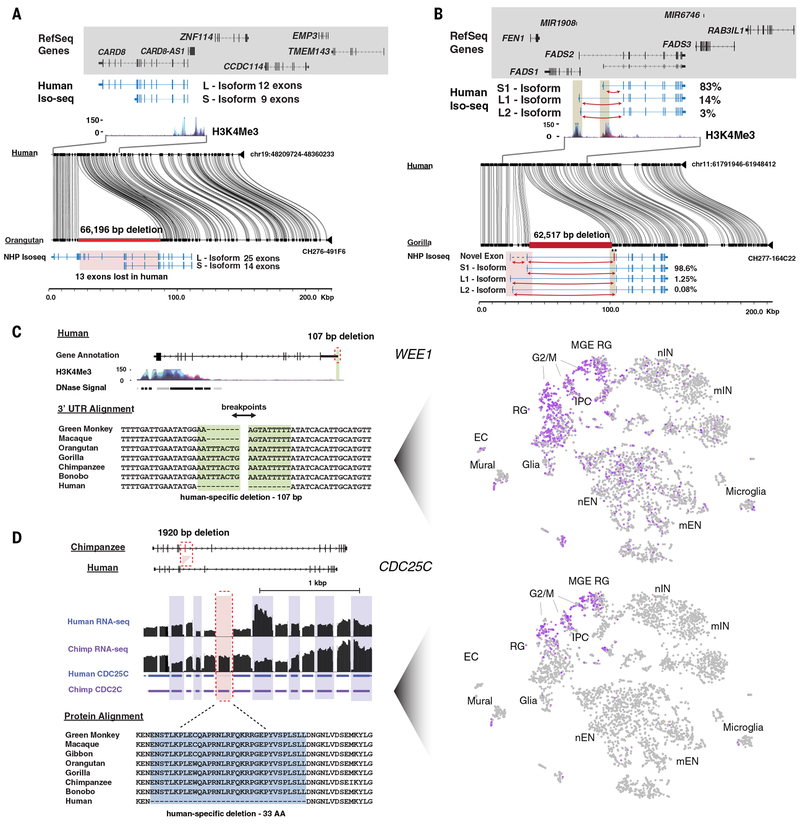
Fig 4. Examples of intragenic human-specific structural variation.
Shown are annotated MSAs between the human reference (GRCh38) and nonhuman primates (NHPs) generated with MAFFT or visualized with Miropeats against sequenced large-insert primate clones. Single-cell gene expression for select genes is highlighted across 4,261 cells developing human telencephalon plotted using t-distributed stochastic neighbor embedding (tSNE) (67). a) A 66.2 kbp intragenic deletion of CARD8 removes 13 putative coding exons in human. Iso-Seq data from chimpanzee and human iPSCs identifies isoforms with and without the deleted exons, respectively. b) A 62.5 kbp intergenic deletion of FADS2 is found in humans, along with an altered isoform ratio: the relative abundance of the long isoforms is increased in humans relative to chimpanzee, as seen in the counts of junction-spanning short reads specific to each isoform. Additionally, a novel, rare (<5%) 75 bp exon is observed in chimpanzee and gorilla but absent in human, likely resulting from a human-specific splice-site mutation. c) A 107 bp deletion in the 3’ UTR of WEE1 reduces AU-rich sequence content in the mRNA. The tSNE plot illustrates that WEE1 is highly expressed in cortical radial glia (RG), intermediate progenitor cells (IPCs), and medial ganglionic eminence progenitors (MGE RG) but shows limited expression in newborn and maturing inhibitory and excitatory neurons (nIN, mIN, nEN, mEN), microglia, endothelial cells (ECs), and glia. d) A 1,920 bp deletion of cell cycle regulator CDC25C removes a 99 bp constitutive exon conserved in mouse, resulting in a 33 amino acid deletion and shorter N-terminal regulatory domain in humans. The tSNE plot illustrates that CDC25C shows restricted expression to telencephalon progenitors in the G2/M cell cycle phase. Human and chimpanzee RNA-seq data were aligned directly to the exonic regions of CDC25C.