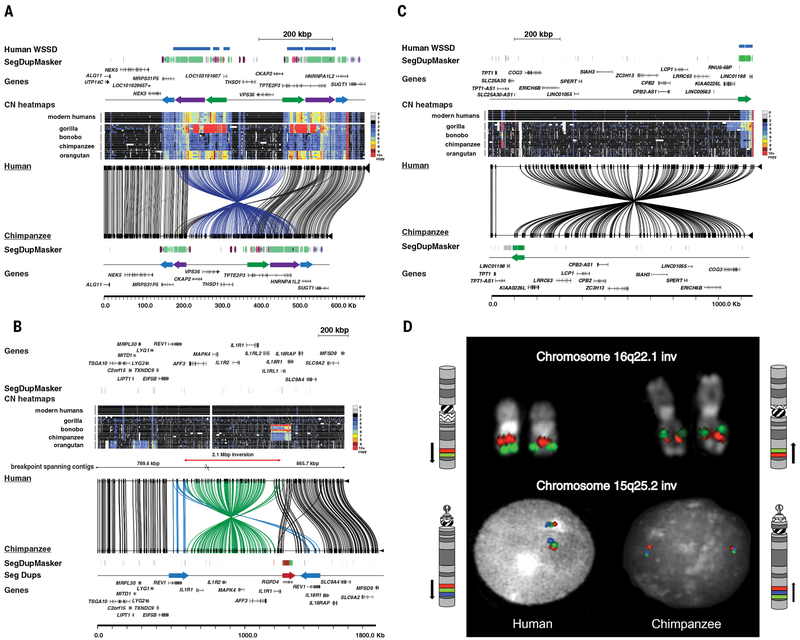
Fig. 5. Complex structural variation.
Large-scale inversions between human and chimpanzee are depicted. The human reference genome sequence (GRCh38) with gene annotation is compared to large-insert clone-based assemblies from the chimpanzee BAC library CH251 using Miropeats. Connecting lines identify homologous regions of high sequence identity. SD organization is depicted as colored arrows as defined by whole-genome shotgun sequence detection (WSSD) and DupMasker. Heatmap indicates copy number (CN) estimated by read-depth from ape genome sequence. a) A ~265 kbp inversion on chromosome 13q14.3 detected by optical mapping in chimpanzee (annotated blue lines). The inverted region is flanked by large ~180 kbp inverted SD blocks that vary with respect to copy number among great apes. b) A 2.7 Mbp inversion on chromosome 2q12-13 detected by BAC end sequencing in chimpanzee (annotated green lines). The inverted region is flanked by duplication blocks containing lineage-specific expansions of the interleukins, an inverted duplication of REV1, and an additional copy of the RGPD4 core duplicon. c) A ~1.1 Mbp inversion at chr13q14.13 identified by optical mapping in chimpanzee encompassing 15 genes. On the telomeric side of the inversion lies a ~60 kbp duplication block that demonstrates lineage-specific duplications in great apes. d) Chromosome inversions, originally detected by optical mapping and BAC end sequencing, confirmed by metaphase analysis and interphase FISH experiments. A human-specific inversion of the chromosome 16q22.1 region was confirmed with orangutan clones CH276-89P20 (red) and CH276-192M7 (green) reported in upper line, and the 15q25.2 inversion was confirmed using chimpanzee clones CH251-321P13 (red), CH251-511D5 (green) and CH251-66E11 (blue) reported in lower line.