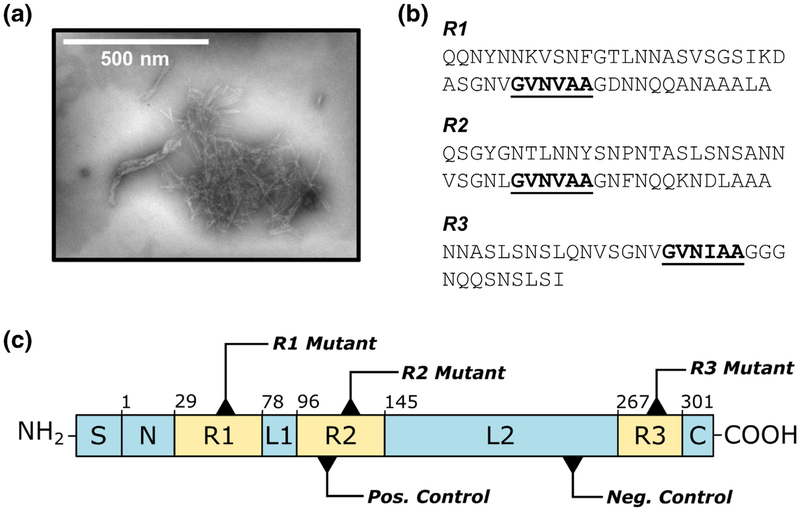
Fig. 1.
The FapC protein from P. aeruginosa. (a) FapC aggregates to form amyloid fibrils. (b) The FapC sequence is characterized by three conserved repeats, R1, R2, and R3, each of which contains a GVNXAA motif indicated by bold/underlined text. (c) Schematic of the PAO1 FapC sequence, including mutation sites for each of the protein variants used in this study. S = signal peptide, N = N-terminal region (residues 1–28), R1–R3 = repeat regions (residues 29–77, 96–144, 267–300); L1–L2 = linker regions (residues 78–95, 145–266); C = C-terminal region (residues 301–316). The first residue of each region is indicated above its corresponding box. The R1, R2, and R3 mutants incorporate GVNXAA→KFDDTK substitution in each of the three repeats, and the TM incorporates this substitution in all three repeats. The negative control (AQGAKD→KFDDTK) consists of a hexapeptide substitution in L2, and the positive control (SANNVS→GVNVAA) inserts a second amyloidogenic hexapeptide in R2.