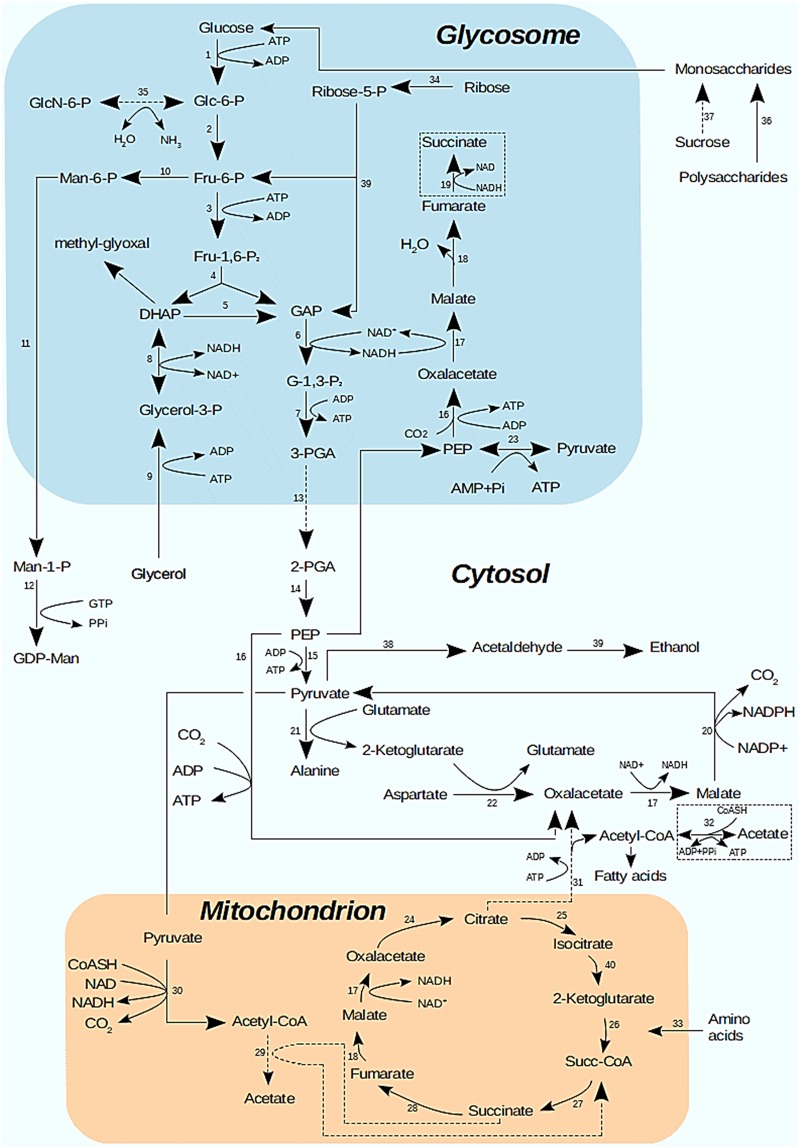
Fig 5. Proposed P. serpens metabolic map based on the present proteomic data as compared to those of Phytomonas sp. EM1 and HART1 [30].
Dashed boxes show enzymes found in the present P. serpens proteome analysis but not in Phytomonas sp EM1 and HART1 genome [30]. Dashed arrows correspond to enzymes found in Phytomonas sp EM1 and HART1 genome but not in the present work. Solid arrows represent enzymes found in P. serpens proteome and in Phytomonas sp EM1 and HART1 genome. Enzymes: 1, hexokinase; 2, glucose-6-phosphate isomerase; 3, 6-phosphofructokinase; 4, fructose-bisphosphate aldolase; 5, triosephosphate isomerase; 6, glyceraldehyde-3-phosphate dehydrogenase; 7, phosphoglycerate kinase; 8, glycerol-3-phosphate dehydrogenase; 9, glycerol kinase; 10, mannose-6-phosphate isomerase; 11, phosphomannomutase; 12, mannose-1-phosphate guanyltransferase, 13, phosphoglycerate mutase; 14, enolase; 15, pyruvate kinase; 16, phosphoenolpyruvate carboxykinase; 17, malate dehydrogenase; 18, fumarate hydratase; 19, NADH-dependent fumarate reductase; 20, malic enzyme; 21, alanine aminotransferase; 22, aspartate aminotransferase; 23, pyruvate phosphate dikinase; 24, citrate synthase; 25, isocitrate dehydrogenase; 26, 2-oxoglutarate dehydrogenase; 27, succinate-CoA ligase; 28, succinate dehydrogenase; 29, acetate: succinate CoA transferase, 30, pyruvate dehydrogenase; 31, citrate lyase; 32, acetyl-CoA synthetase; 33, amino acid oxidation pathway; 34, ribokinase; 35, glucosamine-6-phosphate deaminase; 36, glucoamylase; 37, invertase; 38, pyruvate descarboxylase, 39, alcohol dehydrogenase, 40. isocitrate dehydrogenase.