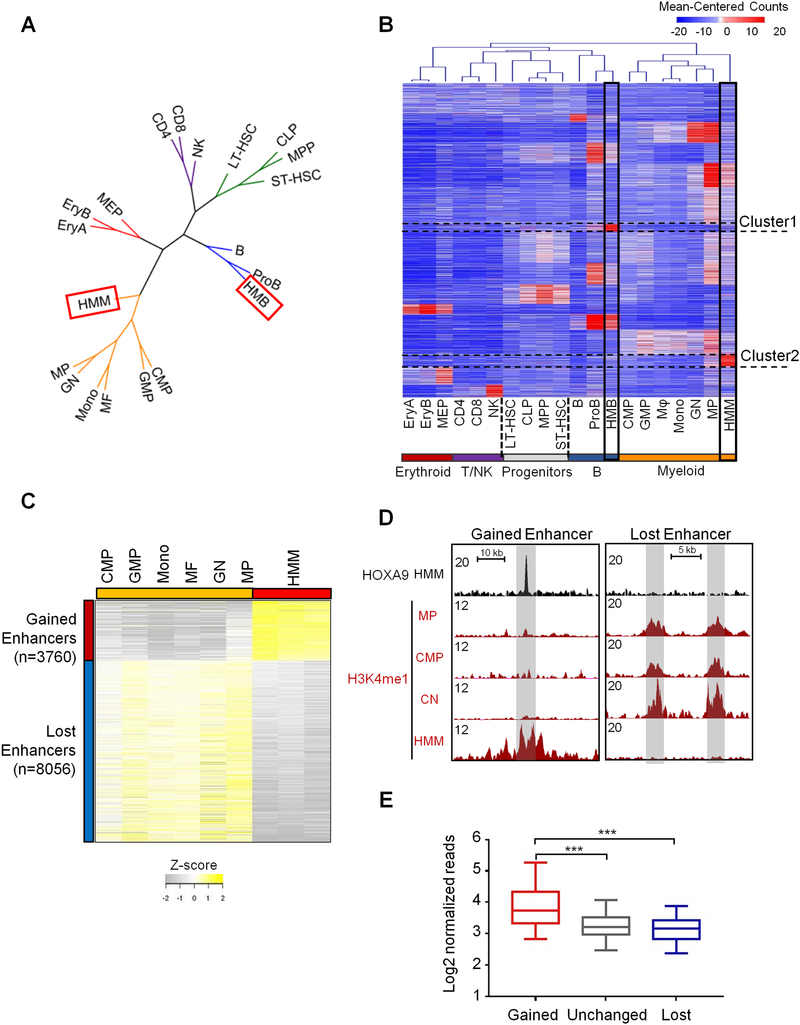
Figure 2. HOXA9/MEIS1-mediated transformation reshapes the enhancer landscape of the leukemia cells.
(A) Clustering dendrogram of cell types based on H3K4me1 profiles to show association of HMM and HMB cells with cells in the myeloid branch and proB cells, respectively. Color code: green for multipotent progenitors, orange for myeloid lineage (including oligopotent common myeloid progenitors (CMP), granulocyte-macrophage (GMP) and differentiated myeloid cells, blue for B lineage, red for erythroid lineage, and purple for T and NK cells. (B) Heat map for 101,413 H3K4me1 marked enhancers for cells as indicated on bottom. H3K4me1 signal was clustered with K-means (K=16) using the normalized read counts at each enhancer region. Cluster 1 and Cluster 2 de novo enhancers were highlighted. (C) Heat map for 11,816 differentially enriched H3K4me1 regions between HMM cells and normal myeloid cells. Cut-off: FDR < 0.05. (D) UCSC browser views of H3K4me1 profiles on representative genomic loci in CMP, GN, MP and HMM cells. Gained (left) and lost (right) enhancers are shaded in grey. (E) Average HOXA9 ChIP-seq tag density at gained, lost and unchanged enhancers as indicated on bottom. Mann-Whitney U-test was used for statistical analysis. Line, median; box, interquartile range (25%–75%); whiskers, 5 and 95 percentiles; ***, p<0.001. See also Figure S2.