Figure 2.
Gfat2 Is an ISC Autonomous Regulator of Cell Division and Cell Growth
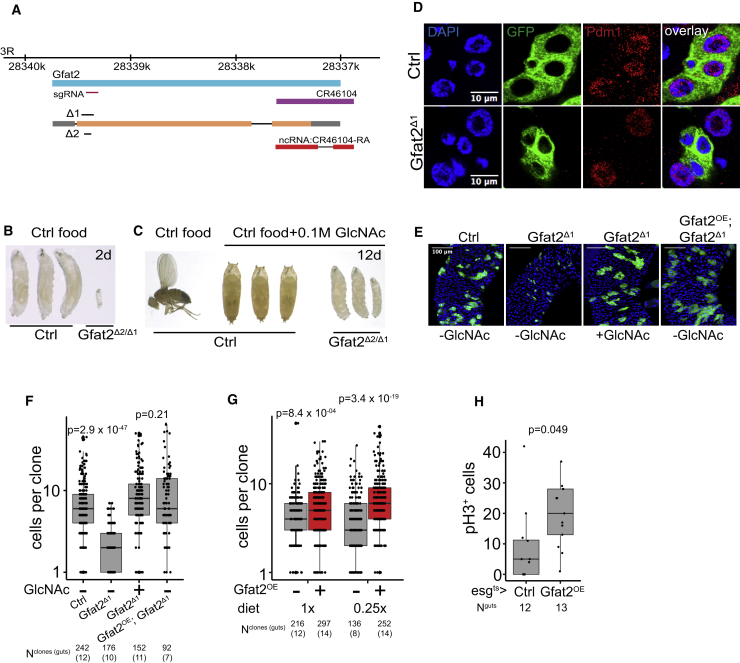
(A) Schematics of the genomic location of the gfat2 gene and the recovered alleles used in this study.
(B) gfat2 null animals are first instar lethal but rescued by dietary GlcNAc. gfat2Δ1/Δ2 trans-heterozygote and wild-type controls 2 days after hatching in control diet.
(C) Wild-type control 12 days after hatching in control diet (adult fly in left) and gfat2Δ1/Δ2 trans-heterozygote and control animals in a diet supplemented with 0.1 M GlcNAc (pupae and larvae on right).
(D) Cells of the gfat2Δ1 intestinal clones are growth defective and lack an enterocyte marker. Control (upper inset) and gfat2Δ1 (lower inset) MARCM clones stained with the enterocyte marker anti-Pdm1 antibody.
(E) Intestinal gfat2Δ1 clones are growth defective and rescued by dietary GlcNAc. MARCM clones of control -GlcNAc, gfat2Δ1 -GlcNAc, gfat2Δ1 +GlcNAc, and UAS-Gfat2; gfat2Δ1 -GlcNAc.
(F) Quantification of (E).
(G) Intestinal MARCM clones overexpressing Gfat2 are larger than controls due to increased cell numbers. Quantification of cell numbers in control and UAS-Gfat2 MARCM clones in the control diet (1×) and calorie-restricted diet (0.25×).
(H) Overexpression of Gfat2 by the Esg-Gal4ts driver leads to an increased midgut mitotic index. Quantification of the pH3-positive cells from Esg-Gal4ts>control and Esg-Gal4ts>UAS-Gfat2 intestines. p values in (F) and (G) are calculated by Wilcoxon rank-sum test with multiple testing correction (FDR < 0.05). p values in (H) are calculated by Wilcoxon rank-sum test. The number of samples in the clonal experiments are indicated in the figure and in Table S3.
See also Figures S1 and S2.