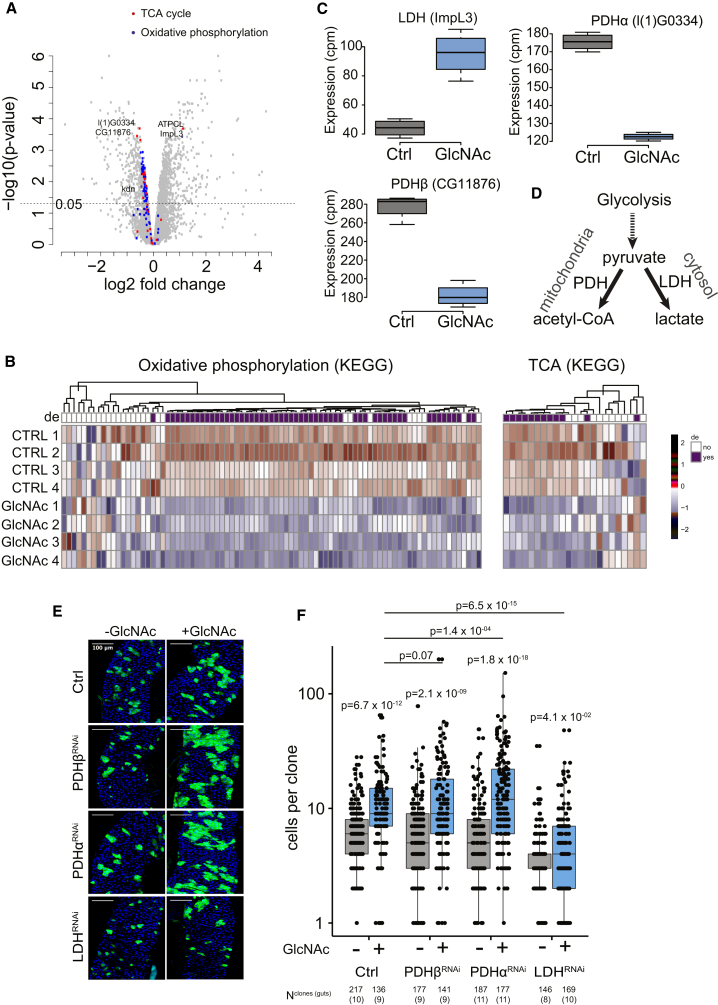
Figure 3.
ISC Activation through HBP Is Mediated by Pyruvate Metabolism
(A) Volcano plot showing the global gene expression changes in midgut cells after GlcNAc feeding. Genes involved in the TCA cycle (KEGG) (red dots) and oxidative phosphorylation (KEGG) (blue dots) are shown.
(B) Heatmaps showing gene expression changes of all expressed genes annotated for TCA cycle (KEGG) and oxidative phosphorylation (KEGG) in midgut cells after GlcNAc feeding. DE sidebar denotes differential expression (violet bar for differentially expressed).
(C) mRNA expressions (counts per million, cpm) of lactate dehydrogenase (LDH) and pyruvate dehydrogenase (PDHα/β) on control versus GlcNAc diet.
(D) Schematics of the role of PDH and LDH in pyruvate metabolism driving pyruvate conversion to acetyl-CoA and lactate, respectively.
(E) Modulating pyruvate metabolism through PDH and LDH knockdowns alters ISC responsiveness to dietary GlcNAc. MARCM clones of control, PDHαRNAi, PDHβRNAi, and LDHRNAi in 0.25× calorie-restricted diet with GlcNAc supplementation.
(F) Quantification of (D). p values in (E) are calculated by Wilcoxon rank-sum test with multiple testing correction (FDR < 0.05). The number of samples in the clonal experiments are indicated in the figure and in Table S3.