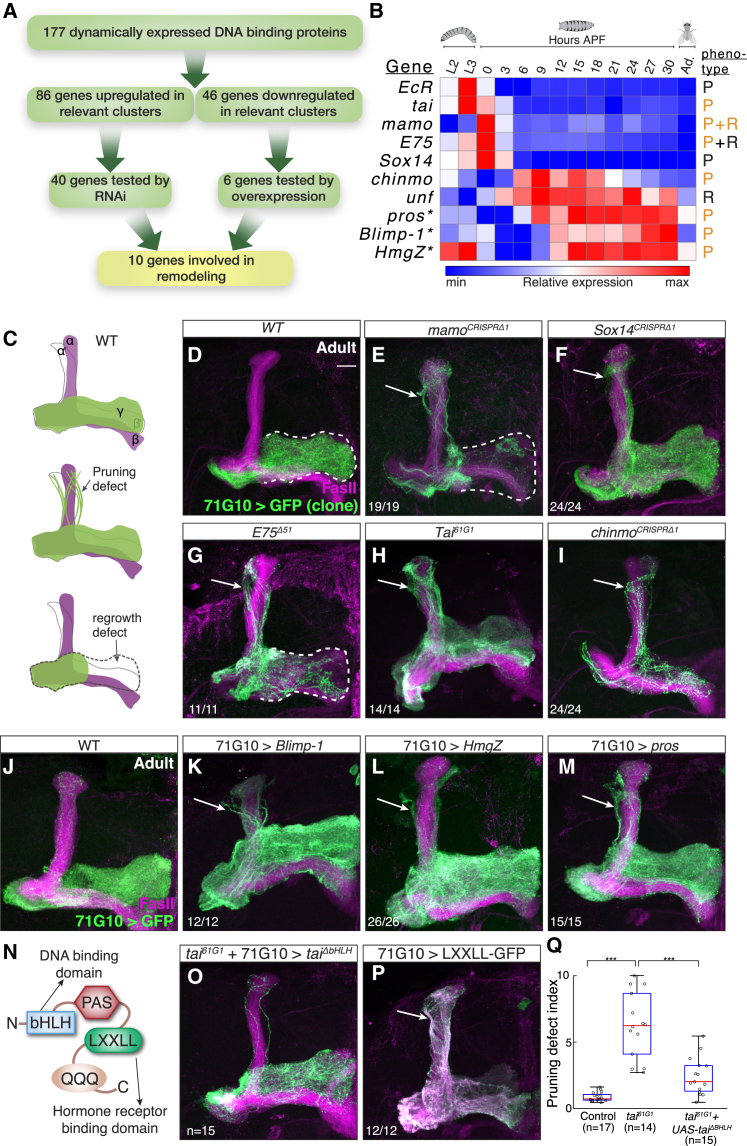
Figure 3.
Dynamic Expression of DNA Binding Proteins Highlight New Genes Required for Remodeling
(A) Scheme of the rationale for the DNA binding protein screen, including the number of genes in each step.
(B) Heatmap showing the relative expression pattern of the 10 positive hits in the DNA binding protein screen. The expression patterns of all 46 genes experimentally tested are shown in Figure S3. Genes were perturbed by RNAi or overexpression (asterisk) experiments. P and R stand for pruning or regrowth phenotypes, respectively. While genes known to be required for remodeling are labeled in black, new findings from this study are labeled in orange.
(C) Schematic representation of WT and defective MBs depicting pruning and regrowth defects. Green represents the γ lobe(s) and magenta represents FasII staining, which in the adult strongly labels α/β neurons, weakly labels γ neurons (not shown for clarity), and does not label αʹ/βʹ neurons.
(D–I) Confocal Z-projections of adult MBs containing WT (D), mamoCRISPRΔ1 (E), Sox14CRISPRΔ1 (F), E75Δ51 (G), Tai61G1 (H), and chinmoCRISPRΔ1 (I) MARCM clones labeled with membrane bound mCD8-GFP (GFP) driven by 71G10-GAL4 (71G10).
(J–M) Confocal Z-projections of adult MBs labeled by 71G10-Gal4 driven mCD8-GFP (GFP) additionally expressing Blimp-1 (K), HmgZ (L), and pros (M).
(N) Schematic diagram of Tai depicting the basic-helix-loop-helix (bHLH) domain required for DNA binding, the LXXLL domain required for binding hormone receptors, the PAS domain, and the poly Q activation domain.
(O) Confocal Z-projections of adult tai61G1 MB MARCM neuroblast clones labeled by 71G10-Gal4 driven mCD8-GFP additionally expressing a tai rescue transgene lacking its bHLH domain (ΔbHLH).
(P) Confocal Z-projection of an adult MB with 71G10-Gal4 driving the expression of mCD8-GFP and as well as the hormone receptor binding domain of Tai fused to GFP.
(Q) Quantification of the pruning severity in (D), (H), and (O). We automatically designated a pruning index to each brain based on image analysis (see STAR Methods). Box centers indicate the median, and the bottom and top edges indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers (99.3% coverage if the data is normally distributed). ∗∗∗p < 0.001; see Figure S6B for a parallel, blind ranking quantification.
Pruning defects, evident by dorsally projecting γ neurons, are marked by arrows, while a regrowth defect, evident by incomplete innervation of the adult γ lobe, is demarcated by a white dashed line. Green is mCD8-GFP driven by 71G10-Gal4; magenta is FasII staining; scale bar represents 15 μm. The numbers (x/n) on the lower left corners depict the number of times the phenotype was observed out of the total hemispheres examined.