Figure 5.
Hierarchical TF Networks Regulate Axon Pruning
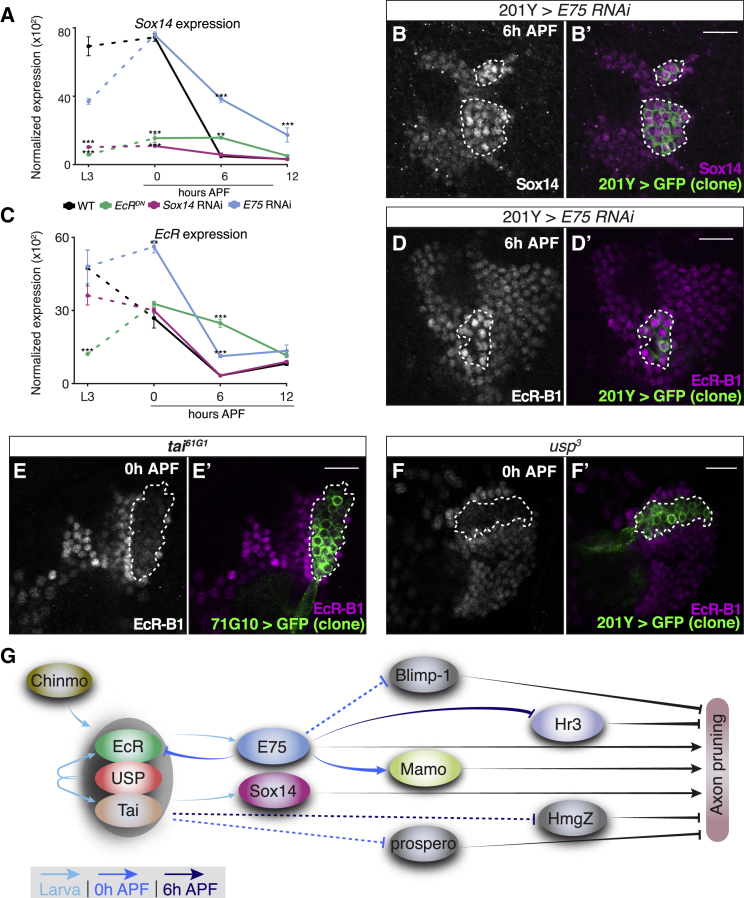
(A and C) Normalized expression of Sox14 (A) or EcR (C) in WT MB γ neurons and in those expressing the indicated transgene (∗∗p < 0.01; ∗∗∗p < 0.001). Error bars indicate SEM; units on the y axis are arbitrary.
(B and D–F) Confocal single slices of the cell body region of MBs containing neuroblast clones labeled with 201Y-GAL4 (B, D, and F) or 71G10-GAL4 (E) driven mCD8-GFP (green), additionally driving the expression of E75 RNAi (B, n = 5; D, n = 4), or mutant for tai61G1 (E, n = 6) or usp3 (F, n = 5). Brains are stained with anti-Sox14 (B) or anti-EcR-B1 (D–F) (magenta) at the indicated time points. Clones are demarcated by dashed lines. The Sox14 antibody staining was increased by 2.2-fold (p < 0.001) within clones expressing E75 RNAi. Expression levels of EcR exhibited a 2.5-fold increase (p < 0.01) within the clone expressing E75 RNAi and a 2.9-fold (p < 0.001) and 3-fold (p < 0.001) decrease within the usp3 and tai61G1 clones, respectively.
Scale bars represent 15 μm.
(G) Schematic model based on data presented here and in Figure S6, describing the hierarchical regulation of axon pruning by regulatory factors as uncovered in this study. The gray ellipse encompassing EcR, Usp, and Tai represents the NR complex. The temporal dimension is represented by the blue color tones of the arrows. While full lines were validated by antibody expressed experiments, dashed lines indicated regulation interpreted only from the RNA-seq data.