Figure 6.
Combining Developmental Clustering with Perturbation Sequencing Is a Powerful Strategy to Reveal Functional Groups of Genes Co-regulated by TF Modules
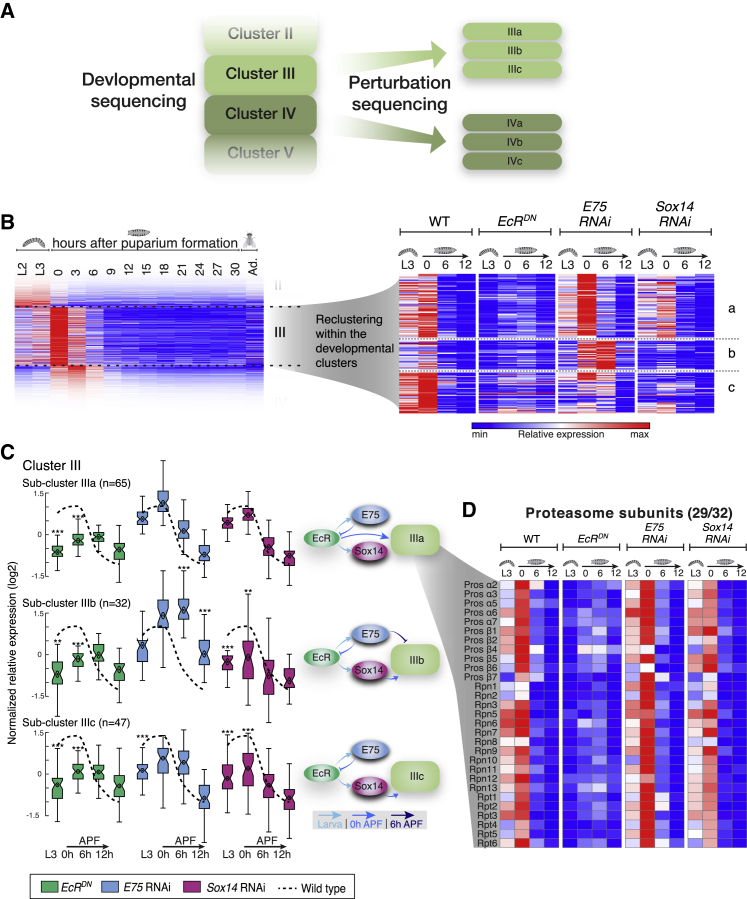
(A) Schematic representation of the sub-cluster analysis. Each developmental cluster was further divided into sub-clusters with a unique pattern of response to the TF perturbations (by k-means clustering; k = 2–4 for each cluster). The sub-clusters contain only genes whose expression significantly changed in at least one perturbation (see STAR Methods for more information).
(B) Heatmaps of developmental cluster III (left) and the three sub-clusters IIIa–c in the perturbation-seq (right) as an example. Similar analyses were conducted for all developmental clusters (see Table S5).
(C) Boxplot analyses of sub-clusters IIIa–c. The average normalized relative gene expression of the WT samples within each sub-cluster is depicted by the dashed line. Boxplots of the normalized relative gene expression for each one of the perturbations in each time point are shown. Box centers indicate the median, and the bottom and top edges indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers (99.3% coverage if the data are normally distributed). Statistical significance was determined (∗∗p < 0.01; ∗∗∗p < 0.001; see STAR Methods) but shown only in cases where the average fold change >2 and thus more likely to also be biologically significant. The hierarchal regulation of each sub-cluster, as inferred from the data, is presented schematically where the temporal dimension is represented by the shades of blue of the arrows.
(D) Heatmap depicting the relative expression patterns of the proteasome subunits, belonging to cluster IIIa, in the different perturbations. Number (x/y) represent the number of genes from the functional group within the sub-cluster (x) / the number of genes from the functional group within the parent cluster (y).