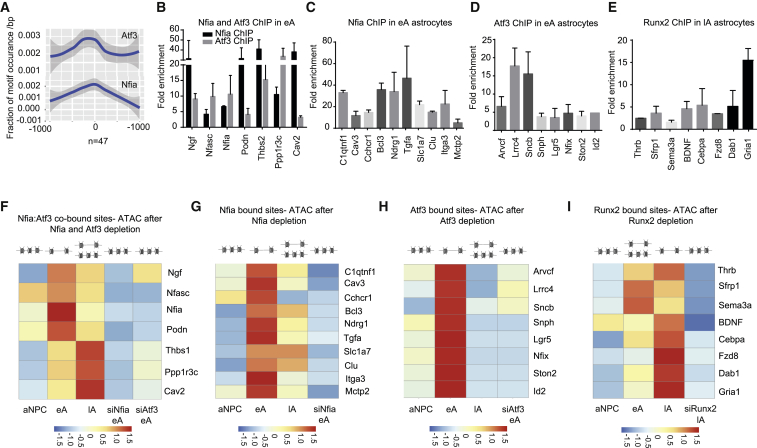
Figure 6.
Nfia, Atf3, and Runx2 Directly Bind Predicted Target Elements and Induce Chromatin Accessibility
(A) Motif enrichment analysis of the set of commonly deregulated and H3K27ac-enriched genes (47 genes) upon Nfia and Atf3 depletion.
(B–E) ChIP-qPCR following Nfia, Atf3, and Runx2 ChIP to detect the Nfia and Atf3 co-occupied (B), Nfia (C), Atf3 (D), and Runx2 (E) binding at predicted Nfia, Atf3, and Runx2 motif sites in the candidate regulatory regions. Enrichments are plotted as a ratio of precipitated DNA (bound) to total input DNA and then further normalized to an intergenic region.
(F–I) qPCR following ATAC in aNPC, eA, and lA and after Atf3 and Nfia (F; eA), Nfia (G; eA), Atf3 (H; eA), and Runx2 (I; lA) depletion at regulatory regions listed in (B)–(E), depicted as a heatmap. Enrichments are plotted as in (B)-(E).