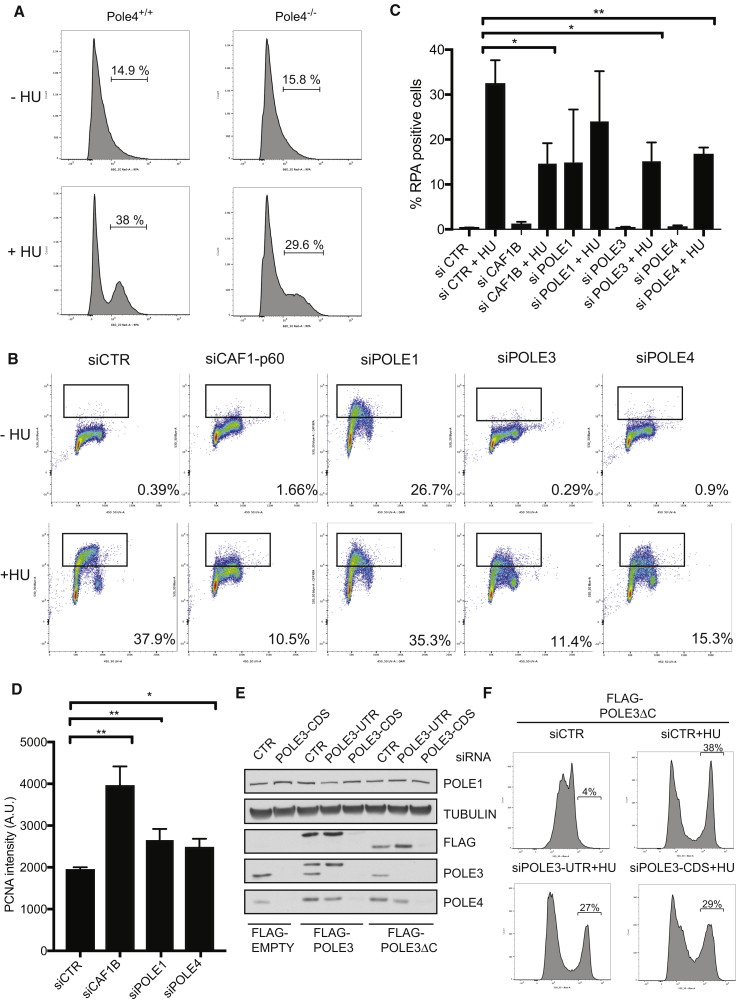
Figure 6.
Constitutive or Transient Depletion of the POLE3-POLE4 Complex Affects Nucleosome Disruption/Maturation at the Replication Fork
(A) FACS analysis of endogenous chromatin-bound RPA from POLE4+/+ and POLE4−/− MEFs treated or not with 2 mM hydroxyurea for 2 hr.
(B) FACS analysis of chromatin from U2OS cells stably expressing GFP-RPA, subjected or not to HU treatment as described in (A). Cells were transfected with siRNAs against the indicated genes, and chromatin purification and FACS analysis were performed 48 hr later.
(C) Bar graph showing percentage of chromatin RPA in cells transfected with the indicated siRNA and treated or not with 2 mM HU for 2 hr. Gates for quantification were selected as shown in (B). Biological triplicates (n = 3) are reported with mean, standard deviation, and p value (∗p < 0.05, ∗∗p < 0.01).
(D) Bar graph showing the median of PCNA staining intensity (arbitrary units) from U2OS RFP-PCNA transiently transfected with siRNAs against the indicated genes. Biological triplicates (n = 3) are reported with mean, standard deviation, and p value (∗p < 0.05, ∗∗p < 0.01).
(E) Western blot analysis of cell lysates from HeLa TRex cells expressing empty FLAG, FLAG-POLE3, or FLAG-POLE3ΔC and transfected with the indicated siRNAs in the presence of doxycycline. After SDS-PAGE and nitrocellulose transfer, membranes were incubated with antibodies against the indicated proteins.
(F) FACS analysis of endogenous chromatin RPA from HeLa TRex FLAG-POLEΔC cells transfected with the indicated siRNAs and treated or not with 2 mM hydroxyurea for 2 hr.