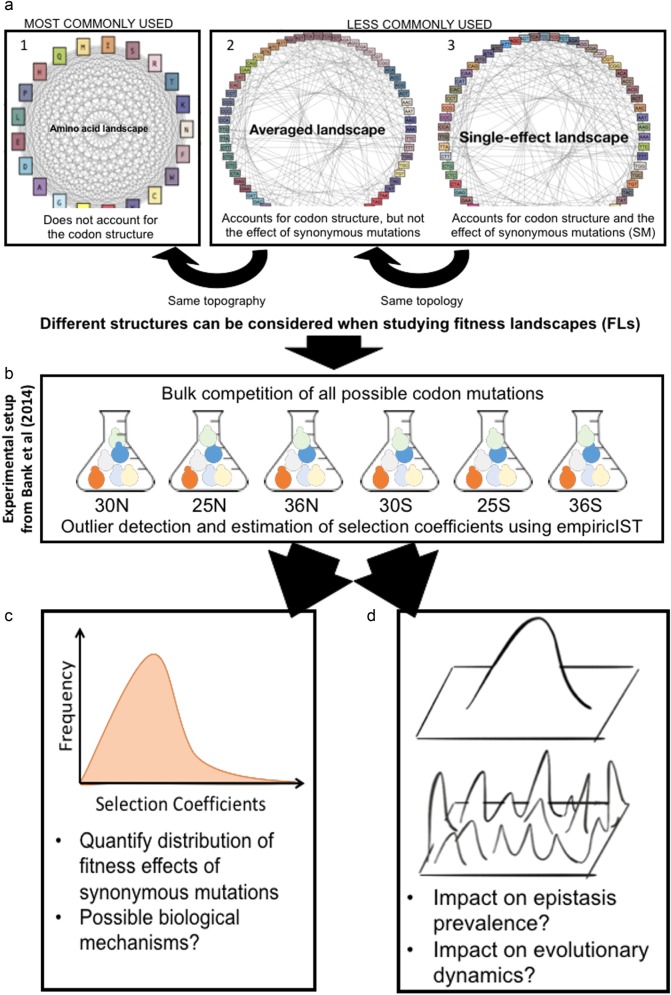
Fig. 1.
Graphical summary of the study. a The consideration of the codon structure and the fitness effects of synonymous mutations results in fitness landscapes with different topologies and topographies. The graphs illustrate fitness landscapes at a single amino acid position. Gray lines indicate single-step mutations and colors indicate potential fitness differences. (1) Many studies implicitly assume that all amino acids are connected by a single mutational step. (2) The codon table restricts the number of possible substitutions at the amino acid level and thus results in a different topology. We denote the fitness landscape that accounts for the codon table but neglects the potential effects of synonymous mutations as the averaged landscape (codons that code for the same amino acid are presented in similar colors). (3) We denote the fitness landscape that considers the individual effect of each codon as the single-effect landscape (each codon has a specific color). b For this study, we obtained deep mutational scanning data of 54 codon fitness landscapes from Bank et al. (2014). We infer individual selection coefficients using a newly developed analysis software, empiricIST. c We quantify the distribution of synonymous fitness effects and perform regression methods to relate these effects to biological mechanisms. d We quantify the shape of the codon fitness landscapes across environments. We illustrate the consequences of ignoring synonymous effects on the evolutionary dynamics on the landscapes