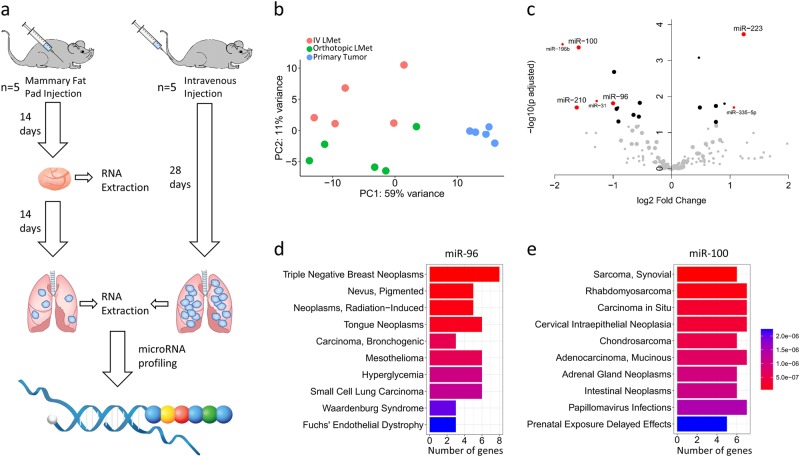
Fig. 1. miRNome analysis of LMets from orthotopic and IV models reveals miR-96 as a potential metastasis suppressor gene.
a Schematic representation of experimental design. Mice were injected with 4T1 cells orthotopically or intravenously. On day 14, primary tumors were excised from the orthotopic group, and RNA purified. On day 28, lungs were excised from mice in both groups, and 1-2 lung macrometastases isolated for RNA extraction and miRNA profiling. b PCA analysis of NanoString miRNA expression in primary tumors (blue), orthotopic LMets (green), and IV LMets (red) shows distinct characterization of each group. The primary tumor samples cluster very tightly compared to the diffuse clustering of the LMet samples. This indicates that as metastatic disease progresses, miRNA profiles become highly variable compared to those of the primary tumor. c. Filtration of significant miRNAs from miRNome analysis is presented as a volcano plot. Each dot represents a different miRNA: large dots represent miRNAs in the top quantile of expression, black dots represent miRNAs with adjusted p-value < 0.05, and red dots represent miRNAs with both a fold change >2 and adjusted p-value < 0.05. MeSH enrichment analysis for d Twenty-one miR-96 and e Twenty miR-100 validated gene-targets obtained from miRTarBase46. miR-96 enrichment results in breast cancer as a top enriched disease. The X-axis represents the number of genes reported to be involved in the diseases labeled in the Y-axis, colors represent adjusted p-values. Barcoded DNA and probe image in (a) was adapted from the NanoString website (https://www.nanostring.com/products/mirna-assays)