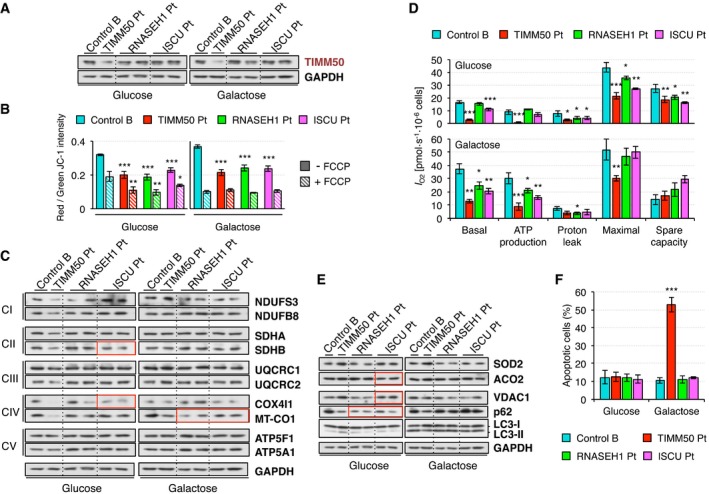
Figure 7. Characterization of RNASEH1 and ISCU mutant fibroblasts.
-
AWestern blot of whole cell extracts for TIMM50 and GAPDH in control and TIMM50, RNASEH1, and ISCU mutant fibroblasts grown in either glucose‐ or galactose‐containing medium. Other components of the protein import machinery are shown in Appendix Fig S8A and their quantification in Appendix Fig S8B.
-
BMitochondrial membrane potential in control and TIMM50, RNASEH1, and ISCU mutant fibroblasts grown in glucose or galactose using JC‐1 staining in untreated or FCCP‐treated conditions. Data are shown as mean ± SD, n = 4 biological replicates; *P < 0.05, **P < 0.01, ***P < 0.001, Student's unpaired two‐tailed t‐test. Exact P‐values are reported in Table EV1.
-
CWestern blot for representatives of all five respiratory complexes in control and TIMM50, RNASEH1, and ISCU mutant fibroblasts grown in glucose or galactose. GAPDH is from the same blot as panel (A). Quantification based on two biological replicates for RNASEH1 and ISCU mutant fibroblasts is shown in Appendix Fig S9. Significant changes in protein levels in RNASEH1 and ISCU mutant fibroblasts compared to controls have been boxed in red.
-
DOxygen consumption measurements in control and TIMM50, RNASEH1, and ISCU mutant fibroblasts grown in glucose or galactose. Values of basal and maximal respiration along with ATP production‐dependent, proton leak respiration, and spare capacity are shown. Data are shown as mean ± SD, n = 4 biological replicates; *P < 0.05, **P < 0.01, ***P < 0.001, Student's unpaired two‐tailed t‐test. Exact P‐values are reported in Table EV1.
-
ESteady‐state levels of ROS‐related proteins SOD2 and ACO2 and mitophagy‐related proteins VDAC1, p62, and LC3 in control and TIMM50, RNASEH1, and ISCU mutant fibroblasts growing in either glucose or galactose were assessed by Western blot using GAPDH as internal control. GAPDH is from the same blot as panel (A). Quantification is shown in Appendix Fig S10. Significant changes in protein levels in RNASEH1 and ISCU mutant fibroblasts compared to controls have been boxed in red.
-
FQuantification of the number of late apoptotic cells in control and TIMM50, RNASEH1, and ISCU mutant fibroblasts grown in glucose or galactose. Data are shown as mean ± SD, n = 4 or more biological replicates; ***P < 0.001, Student's unpaired two‐tailed t‐test. Exact P‐values are reported in Table EV1.
Source data are available online for this figure.
