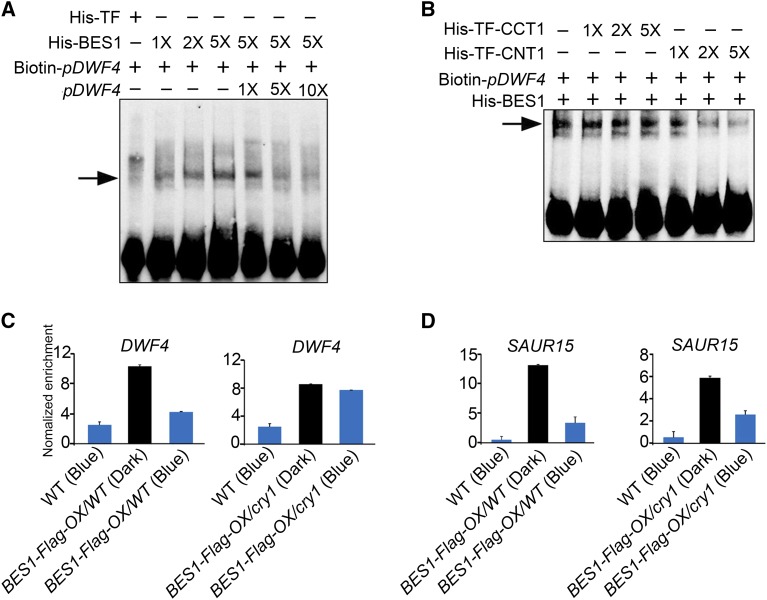
Figure 6.
CRY1 Inhibits the DNA Binding Activity of BES1.
(A) and (B) An EMSA showing that CNT1 inhibits the DNA binding activity of BES1. EMSA was performed with His-BES1 and His-TF proteins using Biotin-pDWF4 as probes. 1×, 2×, 5× and 1×, 5×, 10× in (A) indicate the amount of His-BES1 and cold competitor DNA relative to the initial concentration of His-BES1 and Biotin-pDWF4 probes, respectively. 1×, 2×, and 5× in (B) indicate the amount of His-TF-CCT1 and His-TF-CNT1 relative to that of His-BES1, respectively. Arrows indicate the BES1-DNA complex. Signals at the bottom indicate free probes.
(C) and (D) ChIP-qPCR assay showing that CRY1 inhibits the DNA binding activity of BES1 in Arabidopsis. ChIP-qPCR was performed with BES1-Flag-OX seedlings in the wild type and cry1 mutant background treated with BL for 3 h and dark-adapted or exposed to blue light (100 μmol m−2 s−1) for 1 h. The promoter fragments of DWF4 (C), SAUR15 (D), and TA3 (negative control) were immunoprecipitated with an anti-Flag antibody. IP/input was calculated via a comparison with the Ct values between the immunoprecipitate and input, and relative enrichment was normalized to the negative control TA3. Data are mean values of three biological replicates ± sd.