Figure 2.
Evaluation of Signal Specificity and Evolutionary Conservation of ERD2 Genes in Arabidopsis and N. benthamiana.
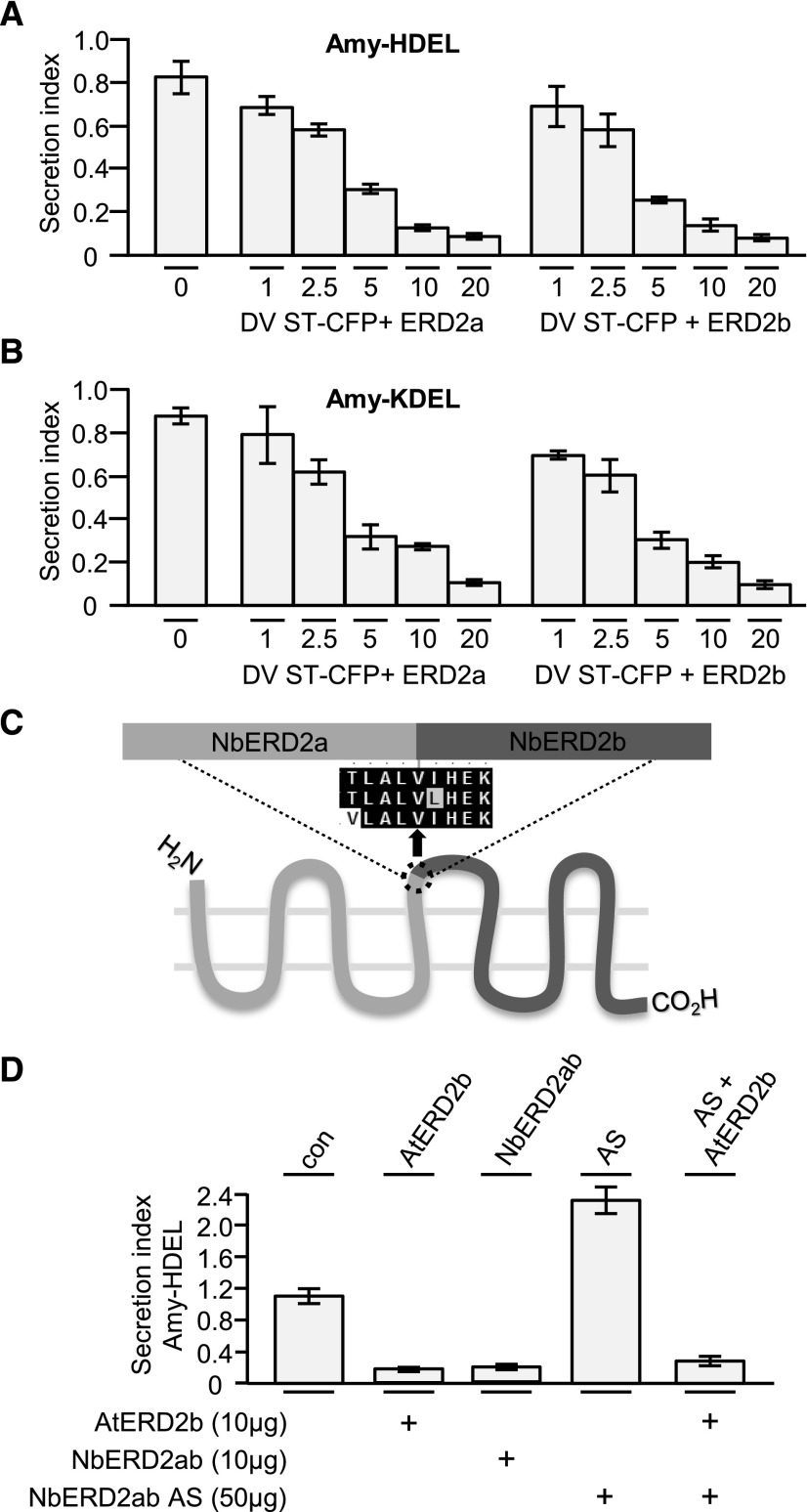
(A) Dose–response assays and experimental setup as in Figure 1E, but comparing ERD2a with ERD2b on Amy-HDEL and using lower amounts of effector plasmids (indicated below each lane in micrograms). Notice the lack of any difference between ERD2a or ERD2b.
(B) Identical experiment as (A), but with Amy-KDEL as cargo instead of Amy-HDEL.
(C) Illustration of the hybrid ERD2 transcript (NbERD2ab), which was generated as sense and as antisense constructs. The alignment shows the point where the fusion was made to generate a hybrid ERD2 coding region.
(D) Transient expression experiment with N. benthamiana protoplasts coexpressing Amy-HDEL with either AtERD2b, sense NbERD2ab, antisense NbERD2ab (AS), or the combination of AS with AtERD2b and incubated for 48 h to allow degradation of endogenous ERD2. Fifty micrograms of cargo plasmid was electroporated alone or coelectroporated together with sense or antisense ERD2 plasmids as indicated by “+.” Error bars are standard deviations of three independent transfections.