Figure 5.
ANAC087 Is Involved in Regulation of Postmortem Nuclear Degradation.
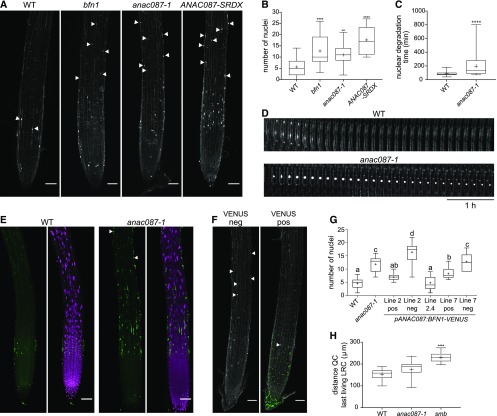
(A) Numerous nuclear remnants were found on the PI-stained root surface of bfn1, anac087-1, and ANAC087-SRDX, as indicated by arrowheads. Fusion of the SRDX domain to a TF turns this TF into a dominant repressor of its target genes (Hiratsu et al., 2003).
(B) Quantification of the number of nuclear remnants shown in (A). Box plots show data from at least 14 roots. Whiskers indicate minimum and maximum values; + indicates average. ANOVA; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
(C) Nuclei of the anac087-1 took longer to be degraded after cell death compared with nuclei from the wild type. Box plots show data from at least 34 nuclei from at least 4 different roots. Whiskers indicate minimum and maximum values; + indicates average. t test; ****P < 0.0001.
(D) Kymograph showing the delay of nuclear degradation in the anac087-1 mutant compared with the wild type. Cells were imaged every 10 min for 5 h.
(E) Multiple TUNEL-positive nuclei were found on the anac087-1 mutant root surface compared with the wild type (indicated by arrowheads). Signal of the fluorescein is shown in green, and DAPI signal is shown in magenta.
(F) Introducing a pANAC087:BFN1-VENUS construct into the anac087-1 mutant resulted in rescue of the mutant phenotype (VENUS pos). In contrast, in segregating VENUS-negative sister plants (VENUS neg), the anac087-1 phenotype was observed. Green signal indicates VENUS; the white signal shows PI staining. Bars in (A), (E), and (F) = 50 µm.
(G) Quantification of the number of nuclear remnants observed in segregating and homozygous pANAC087:BFN1-VENUS lines in the anac087-1 mutant background. Line 2 positive and line 7 positive show a partial rescue of the anac087-1 phenotype. Negative sister plants of the same segregating lines (line 2 neg and line 7 neg) do not show a rescue of the anac087-1 mutant phenotype. Line 2.4 is a homozygous T3 line and shows complete rescue of the anac087-1 mutant phenotype. The experiment was performed on at least 10 roots per genotype. Box plot whiskers indicate minimal and maximal values; + indicates mean. ANOVA; different letters indicate statistically significant differences.
(H) The distance from the QC to the beginning of the last living LRC cell (closest to the transition zone) was altered in the smb mutant, but not in the anac087-1 mutant compared with the wild type, which indicates that timing of LRC PCD is not changed in the anac087-1 mutant. The experiment was performed on at least 13 roots per genotype. Box plot whiskers indicate minimal and maximal values; + indicates mean. Kruskal-Wallis nonparametric test; **P < 0.01, ***P < 0.001, and ****P < 0.0001.