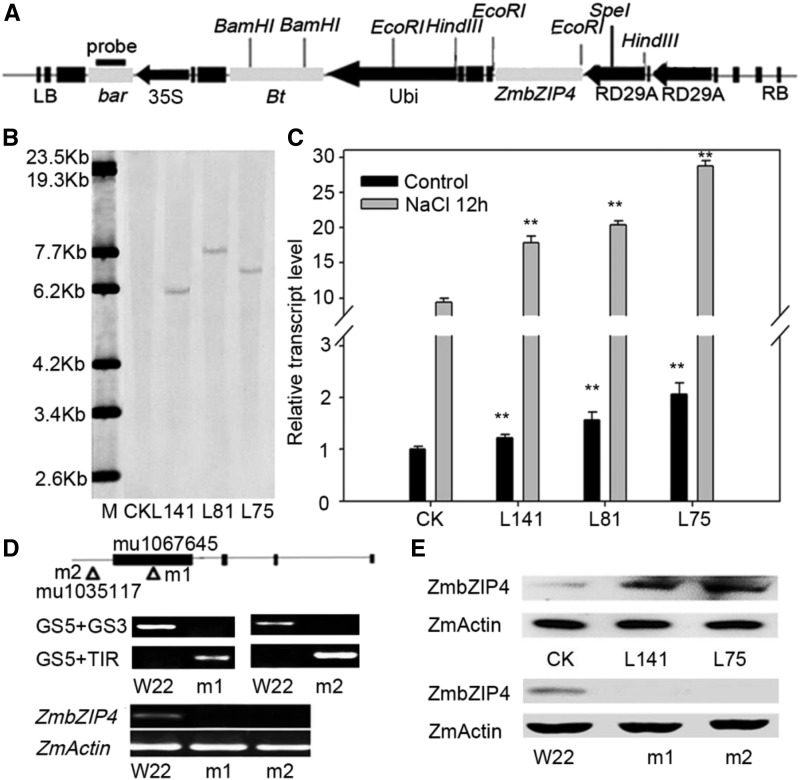
Figure 2.
Molecular identification of the ZmbZIP4 transgenic lines and zmbzip4 mutants. A, T-DNA region of the plasmid of pB7WG2.0-P35S:Bar-Prd29A-Prd29A:ZmbZIP4. The black and gray boxes and black arrows indicate the T-nos, genes, and promoters, respectively. bar, The phosphinothricin acetyltransferase gene; Bt, the cryIAc gene from the Bacillus thuringiensis Ber line; LB and RB, left and right border, respectively; RD29A, the Arabidopsis RD29A promoter; 35S, the cauliflower mosaic virus 35S promoter; Ubi, the maize ubiquitin promoter. B, Southern-blot analysis of the overexpression plants (L141, L81, and L75) and the wild type (CK; DH4866). M, λ-EcoT14 I digest DNA marker. C, Real-time RT-qPCR results of the ZmbZIP4 transgenic genes in both natural and NaCl treatment conditions (100 mm NaCl treatment for 12 h). Bars represent means ± sd (n = 3 repeats). The transcript levels of the ZmbZIP4 overexpression lines were compared with their wild type (CK) under control and NaCl treatment conditions, respectively. Significant differences are indicated by asterisks (Student’s t test: **, P < 0.01). D, ZmbZIP4 UFMu mutants from the maize stock center. Triangles represent the insertion sites, and the molecular identification of the homozygous mutants (m1 and m2) was carried out by PCR and RT-qPCR. E, Western-blot assay of ZmbZIP4 in the transgenic lines and the zmbzip4 mutants.