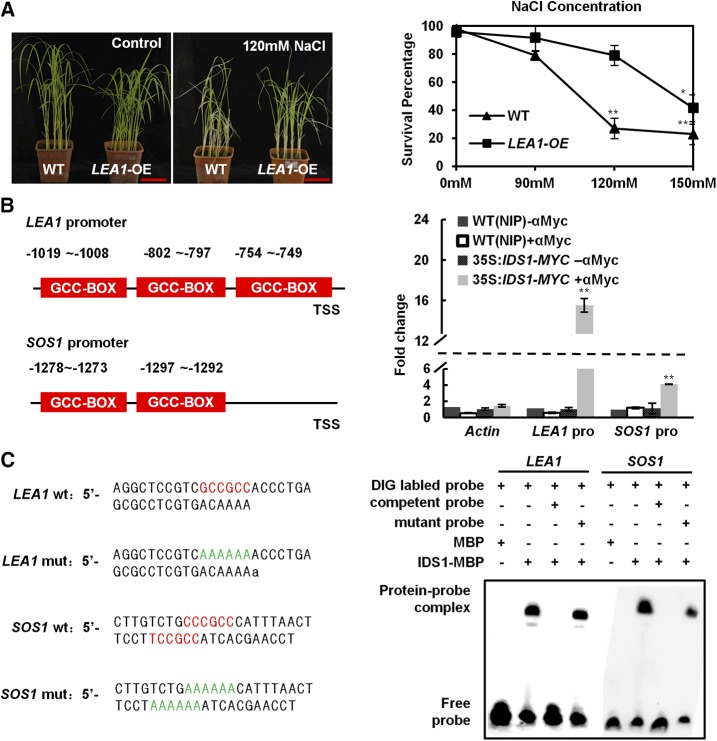
Figure 5.
IDS1 directly binds to the LEA1 promoter. A, Improvement of salt tolerance in LEA1-OE compared to wild-type (WT) seedlings under NaCl stress. Left, salt treatment phenotype; right, the statistics of survival percentage. The seedlings of wild-type and LEA1-OE lines were separately treated with or without (control) NaCl for 10 d (n ≥ 30). Scale bar, 5 cm. Error bars represent sds among three independent replicates. *P < 0.05 and **P < 0.01 (Student's t test). B, ChIP-qPCR assay detected IDS1 enrichment level on the LEA1 and SOS1 promoters. Schemes illustrate the 2-kb promoters of LEA1 and SOS1 (left). The red boxes show the positions of GCC-box motifs. Validation of IDS1 direct binding sites in the LEA1 and SOS1 promoters by ChIP-qPCR analysis (right). Three biological replicates were performed. Error bars represent sds among three independent replicates. **P < 0.01 (Student’s t test). C Direct binding of IDS1 to the LEA1 and SOS1 promoters in EMSAs. The wild-type probes containing GCC-box motifs (red color) were derived from LEA1 and SOS1 promoters, while LEA1 mut and SOS1 mut show probes with mutated GCC-box motifs (AAAAAA, green color). Competition was performed with 125-fold wild-type or mutated cold probes. MBP protein was used as a negative control.