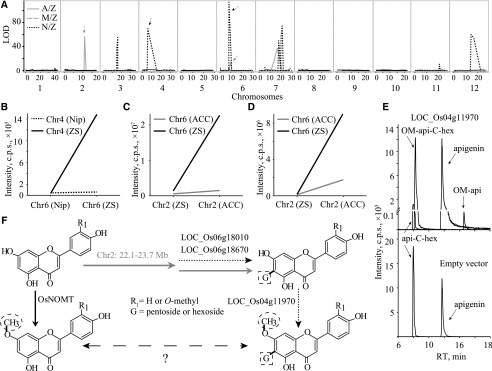
Figure 6.
Pathway dissection for the biosynthesis of O-methylapigenin C-pentoside. A, Mapping result for metabolite m0424 (putative O-methylapigenin C-pentoside) in the three CSSL populations. Vertical gray lines indicate the separation of chromosomes. B, Epistasis analysis of loci on chromosomes 6 and 4 for O-methylapigenin C-pentoside (m0424). C, Epistasis analysis of loci on chromosomes 2 and 6 for metabolite O-methylapigenin C-pentoside (m0424). D, Epistasis analysis of loci on chromosomes 2 and 6 for apigenin 6-C-pentoside (m0400). E, LC-MS chromatograms of in vitro enzyme assays showing the enzyme activity of recombinant LOC_Os04g11970. Protein extract from E. coli containing the pGEX-6p-1 empty vector was used as a negative control. OM-api, O-Methylapigenin; api-C-hex, apigenin C-hexoside; OM-api-C-hex, O-methylapigenin C-hexoside; RT, retention time. F, Predicted pathway from m0164 (standard apigenin) to m0424 (putative O-methylapigenin C-pentoside) based on the epistasis analyses and a previous report (Shimizu et al., 2012). The CSSL populations in which the respective QTLs mapped are indicated with different arrows consistent with those in A: gray solid arrows for A/Z and black dashed arrows for N/Z populations.