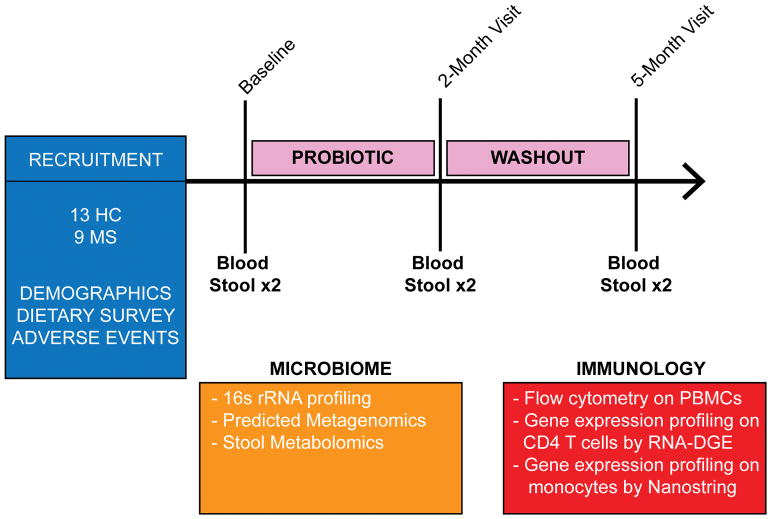
Figure 1. Study design flowchart.
Blood and fecal samples were collected from healthy subjects (n=13) and multiple sclerosis (MS) patients (n=9) prior to (baseline), at discontinuation of probiotic LBS (2-month visit) and 3 months thereafter (5-month visit). Bacteria DNA was extracted from feces samples obtained at all 3 time points and 16S rRNA sequencing was performed using Illumina MiSeq.16S data was used to obtain predicted metagenomics by PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States). Feces samples were also used for stool metabolomics profiling at all 3 time points. Immune cells profiling was performed in healthy subjects and MS patients at all 3 time points by flow cytometry. Gene expression profiling was performed on peripheral monocytes and CD4 T cells from healthy controls and MS patients using a Nanostring immunology panel array and high-throughput eukaryotic digital gene expression RNA-Seq (RNA-DGE) respectively.