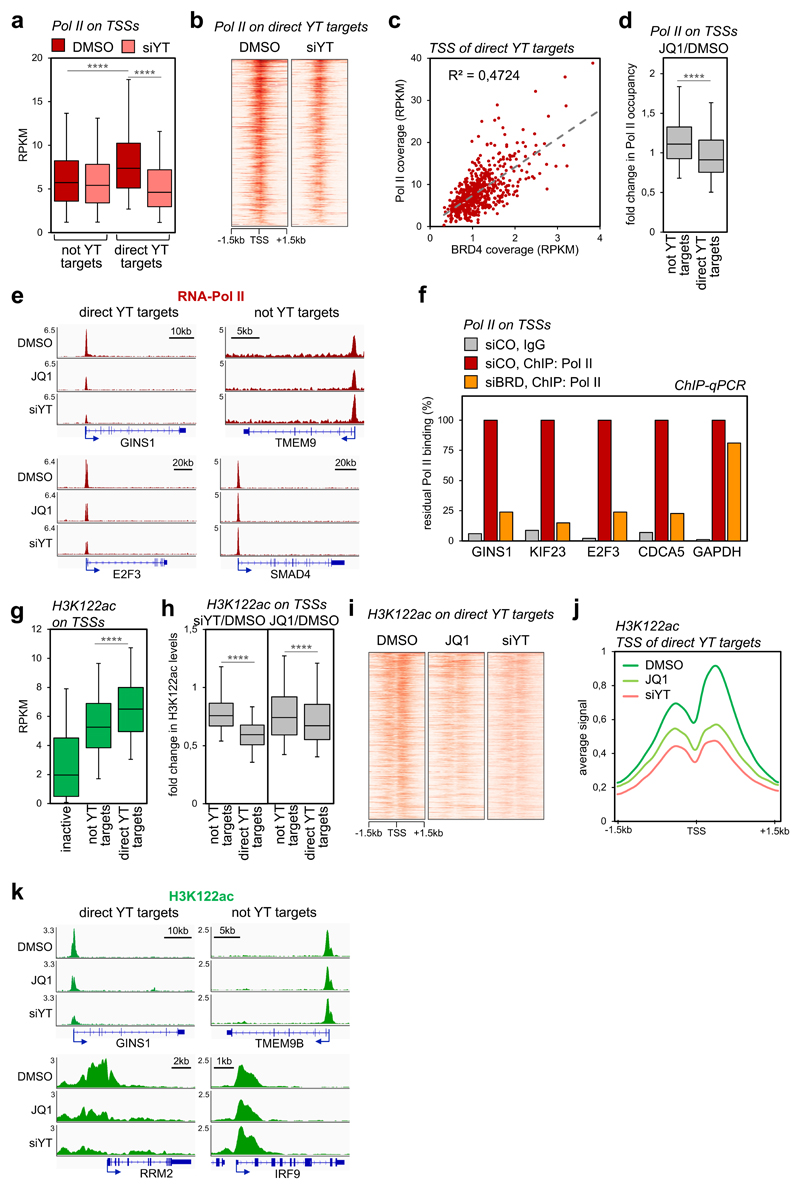
Figure 4. YAP/TAZ and BRD4 regulate Pol II loading and H3K122 acetylation on TSSs.
a) Box-and whiskers plots showing the distribution of RNA-Pol II ChIP-seq signal (expressed as normalized read density, RPKM) comparing promoters of genes not activated by YAP/TAZ (n=8026) or of YAP/TAZ target genes (n=616) in control (DMSO) or YAP/TAZ depleted cells. The box includes values within the 25th and 75th percentile (with the median highlighted by the line in the middle) and whiskers extend from the 5th to the 95th percentile. **** p<10-10 (one-tailed Mann-Whitney U test)
b) Heatmap showing RNA-Pol II loading on the promoters of YAP/TAZ targets in MDA-MB-231 cells, in a window of ±1.5kb centered on the transcription start site (TSS).
c) Linear correlation between BRD4 and RNA-Pol II occupancy (both expressed in RPKM) on the TSS of YAP/TAZ target genes (n=616). r2 was calculated using linear regression analysis (F-test p-value<0.0001).
d) Box-and whiskers plots (defined as in a) showing the change in RNA-Pol II promoter occupancy in JQ1-treated cells vs. control cells (DMSO), comparing promoters of genes not activated by YAP/TAZ (n=8026) with promoters of YAP/TAZ target genes (n=616). **** p<10-10 (one-tailed Mann-Whitney U test)
e) Genome browser view of RNA-Pol II binding profiles at representative promoters of YAP/TAZ target genes or not-YAP/TAZ targets. Pol II binding is reduced upon JQ1 treatment or YAP/TAZ depletion on the TSS of YAP/TAZ targets.
f) ChIP-qPCR verifying RNA-Pol II binding to promoters of established YAP/TAZ targets upon depletion of BET proteins. GAPDH promoter represents a non-YAP/TAZ target. ChIP with pre-immune IgG displayed background signal (which was comparable in all samples). DNA enrichment was calculated as fraction of input and is presented as % of RNA-Pol II binding in control cells (siCO).
g) Box-and whiskers plots of H3K122ac ChIP-seq signal (RPKM) showing its enrichment on YAP/TAZ target genes (n=616) in comparison with inactive promoters (n=4618) and not-YAP/TAZ targets (n=8026). Data are presented as in a. **** p<10-10 (one-tailed Mann-Whitney U test)
h) Box-and whiskers plots showing the change in H3K122ac promoter levels in YAP/TAZ-depleted (left) or JQ1-treated cells (right), both compared to control cells (DMSO), showing preferential loss of H3K122ac at promoters of YAP/TAZ target genes (n=616) vs. promoters of not YAP/TAZ targets (n=8026). Data are presented as in a. **** p<10-10 (one-tailed Mann-Whitney U test)
i) Heat map showing acetylation of H3K122 on the promoters of YAP/TAZ targets in MDA-MB-231 cells, in a window of ±1.5kb centered on the transcription start site (TSS).
j) Average ChIP-seq profile of H3K122ac on the promoters of YAP/TAZ target genes (n=616) in MDA-MB-231 cells, in a window of ±1.5 kb centered on TSS.
k) Genome browser view of H3K122ac levels at representative promoters of YAP/TAZ target genes vs. not-YAP/TAZ targets.